The GRAS-domain transcription factor PAT1 interacts with INDETERMINATE-DOMAIN 3 protein to activate LIPOXYGENASE 3, leading to enhanced JA accumulation and improved cold tolerance in grape.
Abstract
Cultivated grapevine (Vitis) is a highly valued horticultural crop, and cold stress affects its growth and productivity. Wild Amur grape (Vitis amurensis) PAT1 (Phytochrome A signal transduction 1, VaPAT1) is induced by low temperature, and ectopic expression of VaPAT1 enhances cold tolerance in Arabidopsis (Arabidopsis thaliana). However, little is known about the molecular mechanism of VaPAT1 during the cold stress response in grapevine. Here, we confirmed the overexpression of VaPAT1 in transformed grape calli enhanced cold tolerance. Yeast two-hybrid and bimolecular fluorescence complementation assays highlighted an interaction between VaPAT1 with INDETERMINATE-DOMAIN 3 (VaIDD3). A role of VaIDD3 in cold tolerance was also indicated. Transcriptome analysis revealed VaPAT1 and VaIDD3 overexpression and cold treatment coordinately modulate the expression of stress-related genes including lipoxygenase 3 (LOX3), a gene encoding a key jasmonate biosynthesis enzyme. Co-expression network analysis indicated LOX3 might be a downstream target of VaPAT1. Both electrophoretic mobility shift and dual luciferase reporter assays showed the VaPAT1-IDD3 complex binds to the IDD-box (AGACAAA) in the VaLOX3 promoter to activate its expression. Overexpression of both VaPAT1 and VaIDD3 increased the transcription of VaLOX3 and JA levels in transgenic grape calli. Conversely, VaPAT1-SRDX (dominant repression) and CRISPR/Cas9-mediated mutagenesis of PAT1-ED causing the loss of the C-terminus in grape calli dramatically prohibited the accumulation of VaLOX3 and JA levels during cold treatment. Together, these findings point to a pivotal role of VaPAT1 in the cold stress response in grape by regulating JA biosynthesis.
Introduction
Transcription factors (TFs) are proteins that bind to specific DNA motifs in the promoter regions of target genes, thereby regulating their transcription. In plants, TFs play pivotal roles in signaling and regulatory networks in many development processes and stress responses (Xiong et al., 2002; Lindemose et al., 2013; Zhu, 2016). GRAS proteins are one of the many plant TFs families whose name is derived from the first three identified members named gibberellin insensitive (GAI), repressor of GAI (RGA) and scarecrow (SCR). The GRAS TF family consists of eight well-known subfamilies (i.e. DELLA, HAM, LISCL, PAT, LAS, SCR, SHR, and SCL3), many of which were found to be involved in gibberellic acid (GA) regulation and various biological processes in plants (Bolle, 2004; Sun et al., 2012; Richards et al., 2015). Recent studies have now reported the participation of GRAS proteins in stress response in several species. For instance, overexpression of SlGRAS40 improved resistance to drought and salinity via enhanced ability to scavenge ROS (Liu et al., 2017). Most GRAS members lack a DNA-binding domain (BD). For example, DELLA proteins function as a transcriptional activator through the DNA binding of the INDETERMINATE DOMAIN (IDD) family proteins (Yoshida et al., 2014; Hirano et al., 2017; Hakoshima, 2018). Furthermore, DELLAs prevent inhibitory JASMONATE ZIM-domain 1 (JAZ1) interaction with a key transcriptional activator of JA responses, MYC2, and, thus, enhance the ability of MYC2 to regulate its target genes (Hou et al., 2010). However, the underlying mechanisms of many GRAS proteins in stress response are still largely unknown.
In plants, jasmonate acid (JA) plays critical roles in the response to abiotic stress including cold (Wasternack and Hause, 2013; Chini et al., 2016; Hu et al., 2017; Howe, 2018; Wasternack and Feussner, 2018; Yang et al., 2019). Among the various enzymatic proteins involved in JA biosynthetic pathways in plants (Wasternack and Feussner, 2018), oxygenation of a-LeA at the C-13 position by lipoxygenase (LOX) enzymes is the key first step in JA biosynthesis. Recent studies revealed that many 13-LOX contribute to JA formation (Feussner and Wasternack, 2002; Wasternack and Feussner, 2018). In low-temperature conditions, the expression of many JA synthesis-related genes was induced and bioactive JA–Ile is synthesized (Zhu, 2016; Hu et al., 2017). Next, the ICE-CBF transcriptional regulation cascade signaling pathway is activated, and the expressions of cold-regulated genes are induced to improve plant cold tolerance (Yang et al., 2019). Although extensive studies revealed detailed molecular roles of JA in the ICE-CBF pathway and stress responses (Hu et al., 2013), recent findings have indicated that ICE1 may not be an indispensable master regulator of the CBF/DREB1 genes (Thomashow and Torii, 2020). Therefore, the factors responsible for the activation of jasmonate synthesis during cold stress remain unclear and more complex than previously thought.
Low-temperature stress is a major determinant of plant growth and development, and significantly constrains the geographic distribution of plants. Grapevine is a widely cultivated fruit crop worldwide and the most commonly cultivated varieties are mainly derived from Vitis vinifera. Extremely low temperatures in winter can cause serious damage to its buds and branch tissues. In contrast, the wild Amur grape (Vitis amurensis) is extremely cold tolerant, withstanding temperatures as low as −30°C (Fennell, 2004). The genes belonging to the classic cold stress signaling pathway such as the ICE-CBF/DREB1 regulatory pathway in V. amurensis such as VaICE1 and VaICE2 (Xu et al., 2014) and VaCBF4 (Li et al., 2013) have been identified. Moreover, the function of several TFs such as VaERF057 (Sun et al., 2016), VaAQUILO (Sun et al., 2018), VaWRKY33, and VaERF092 (Sun et al., 2019) during cold stress response in V. amurensis were also identified. These results provide pivotal clues for further elucidation of the cold stress response regulatory networks in V. amurensis.
PAT1 is a member of the GRAS TF family and its transcription increased significantly under low temperature treatment in grapevines (Xin et al., 2013). VaPAT1 was also found to be induced by drought and high salinity treatments in V. amurensis. Overexpression of VaPAT1 enhanced cold tolerance in transgenic Arabidopsis (Arabidopsis thaliana), which indicates a functional role in the cold stress response (Yuan et al., 2016). However, exactly how VaPAT1 regulates the downstream genes and contributes to cold tolerance in grapevines remains to be ascertained. In this study, we demonstrate that VaPAT1 improves cold tolerance of grape calli by regulating JA biosynthesis. VaLOX3 was identified as a possible downstream target of VaPAT1 through RNA-Seq and co-expression network analysis. Further experiments indicate that VaPAT1 activates the expression of VaLOX3 by physically interacting with an INDETERMINATE-DOMAIN 3 protein, VaIDD3. Collectively, our data demonstrate that VaPAT1 promotes the JA biosynthesis and enhances cold resistance in grape, providing new insights into the roles of GRAS TFs in regulating abiotic stress resistance in plants.
Results
Overexpression of VaPAT1 improves cold tolerance of grape calli
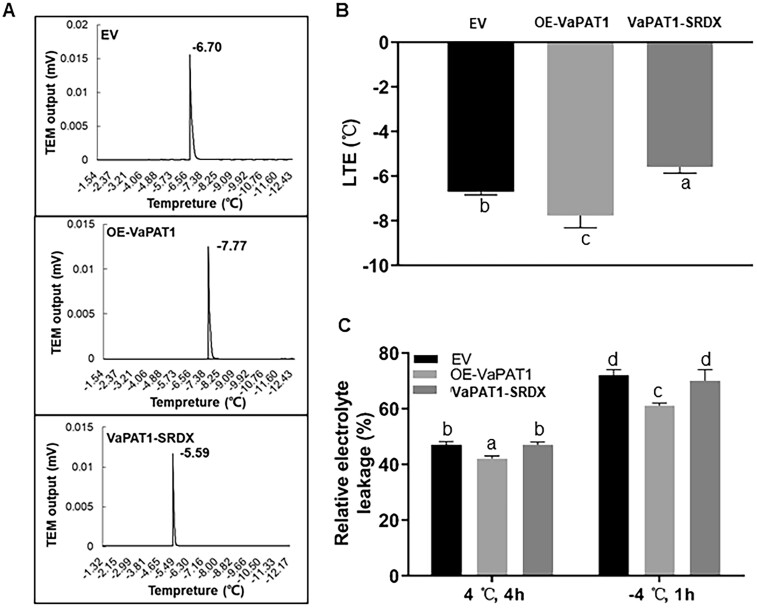
Previous work has shown that overexpression of VaPAT1 conferred cold stress tolerance in transgenic Arabidopsis (Yuan et al., 2016). To validate the function of VaPAT1 in grapevine, the grape calli 41B from Lecourieux et al. (2010) was used to generate transgenic lines. A total of three different transgenic lines were generated which includes the overexpression of VaPAT1 (OE-VaPAT1), suppression of VaPAT1 (VaPAT1-SRDX, based on chimeric repressor gene silencing technology; Mitsuda et al., 2011), and empty vector (EV) as control (Supplemental Figure S1, A). The positive calli were selected by kanamycin (Supplemental Figure S1, B and Supplemental Table S1) and the expression of VaPAT1 and NPTII at the DNA and mRNA levels was confirmed in three independent OE-VaPAT1 and VaPAT1-SRDX transgenic lines (Supplemental Figure S1, C). RT-qPCR analysis showed that VaPAT1 expression was up-regulated in these transgenic calli lines, with a consistent fold change (FC) increase of approximately three-fold when compared with the EV line. These calli were used for cold tolerance evaluation using several key physiological indices including low-temperature exotherms (LTEs) and relative electrolyte leakage (Supplemental Figure S1). The LTEs of the EV, OE-VaPAT1, and VaPAT1-SRDX grape calli were −6.70°C, −7.77°C, and −5.59°C, respectively (Figure 1, A). Significantly lower (−1.07°C) and higher (+1.11°C) LTE in OE-VaPAT1 and VaPAT1-SRDX were observed when compared with the EV control (Figure 1, B). OE-VaPAT1 lines showed significantly lower electrolyte leakage compared with EV transgenic lines at cold (4°C) and freezing (−4°C) temperatures while VaPAT1-SRDX were comparable with EV lines (Figure 1, B). These results indicated that VaPAT1 also positively regulates the cold stress response in grapevines.
Figure 1.
Overexpression of VaPAT1 enhances cold tolerance in transgenic grape calli. A, Cold tolerance evaluation system of grapevine calli. After placing the calli on the thermoelectric modules (TEMs), exotherms were identified manually from a plot of thermistor output (x-axis) versus loaded TEM output minus the empty TEMs output (y-axis). The LTEs (indicating the cell freeze temperatures) were obtained to evaluate the cold tolerance. B, LTEs of transgenic grapevine calli. C, Electrolyte leakage of each line treated at 4°C, 4 h and −4°C, 1 h, respectively. The error bars indicate the SD from triplicate technical repeats. The letters above the bars indicate significant differences (P < 0.05, according to Duncan’s multiple range test).
VaPAT1 localizes to the nucleus
The nuclear localization signal (NLS) is common to the PAT1 subfamily of GRAS TF, however, in cases where the proteins are localized only in the cytoplasm (e.g. AtPAT1) or both the cytoplasm and nucleus (e.g. AtSCL13) has also been documented (Sun et al., 2012). In silico analysis using NLS-mapper, LocSigdb, and NLSdb tools indicated the presence of a partial NLS in the N-terminus of VaPAT1. To ascertain its localization, the ORF of VaPAT1 was fused to the N-terminus of the GFP protein under the control of CaMV 35S promoter (35S:VaPAT1-GFP). The 35S:VaPAT1-GFP and a control GFP construct were independently transformed in Nicotiana benthamiana. VaPAT1-GFP green fluorescence was detected only in the nucleus while the GFP signal was evenly distributed in both the cytoplasm and nucleus (Supplemental Figure S2). This result suggests that VaPAT1 localized in the nucleus and may function as a transcriptional regulator.
VaPAT1 interacts with VaIDD3
GRAS TFs often lack the DNA-BD (e.g. DELLA proteins), therefore, interaction with other intermediate protein(s) is required for DNA binding and transcriptional regulation of downstream target genes (Yoshida et al., 2014; Hirano et al., 2017; Hakoshima, 2018). In Arabidopsis, several INDETERMINATE DOMAIN (IDD) proteins were shown to interact with DELLA proteins to regulate downstream target genes (e.g. SCARECROW-LIKE3, a key GA-positive regulator; Fukazawa et al., 2014). Large-scale yeast two-hybrid (Y2H) screens in Arabidopsis revealed that only AtIDD3 (one of the 16 IDDs) interacted with all known DELLA proteins such as RGA, GAI, and RGL1–3 (Yoshida et al., 2014). Thus, we hypothesized that VaPAT1 may also interact with the homologs of AtIDD3 in grape.
Reciprocal best BLAST identified 15 IDD candidates from the V. vinifera cv “Pinot Noir” (PN40024) genome using 16 A. thaliana IDDs as query (Supplemental Table S2). In particular, VvIDD3 was the predicted ortholog of AtIDD3. Querying PlantTFDB v5.0 database also corroborated our findings. The CDS of IDD3 was cloned from both V. vinifera Muscat Hamburg and V. amurensis, of which both were 100% identical at the DNA sequence and amino acid level in the two species (Supplemental Figure S3). As such, VaIDD3 was used for illustration and downstream analysis here on in.
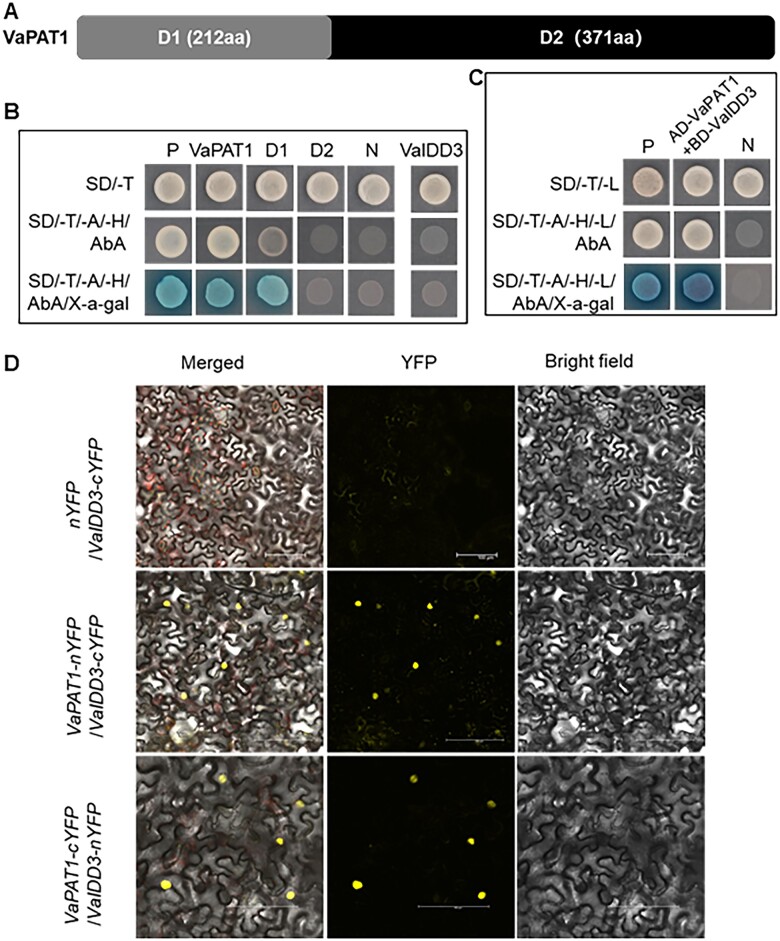
The transcriptional activity of VaPAT1 and VaIDD3 were further tested in yeast (Figure 2, A and B). VaPAT1 was divided into two parts according to the N-terminal and conserved C-terminal (Figure 2, A). The full-length of VaPAT1 and two truncated forms—D1 (aa: 1–212) and D2 (aa: 213–583) were fused to the GAL4 DNA- BD, respectively. The result demonstrated that VaPAT1 and VaPAT1-N term promoted yeast growth on SD/-T/-A/-H plate with X-gal activity (Figure 2, B). VaPAT1-C and VaIDD3 did not grow in the same plates. These data support that VaPAT1 may function as a transcriptional activator.
Figure 2.
VaPAT1 interacts with VaIDD3. A, VaPAT1 protein structural representation in (B). B, Analysis of VaPAT1 and VaIDD3 transactivation activity. VaPAT1 (full length), D1 (N-terminal only) and D2 (C-terminal only), each fused with GAL4 DNA-BD protein were expressed in yeast. The transactivation activity was revealed through the expression of the lacZ reporter gene (β-galactosidase activity). Vectors pGBKT7 and pGBKT7-53 + pGADT7-Rec2-53 were expressed in yeast as a negative (N) and a positive (P) control, respectively. C, Y2H assay showing VaPAT1 and VaIDD3 interaction. P and N indicate the positive and negative control, respectively. D, In vivo interactions between VaPAT1 and VaIDD3 proteins by BiFC assays in N. benthamiana leaf cells. The reconstitution of YFP is shown. Scale bars, 100 μm.
To test the potential interaction between VaIDD3 and VaPAT1, Y2H assay was employed. As bait, the full-length CDS of VaIDD3 was fused to the GAL4 DNA-BD (BD-VaIDD3) while the full-length CDS of VaPAT1 fused to the GAL4 DNA-activation domain (AD-VaPAT1) as prey. In the Y2H assay, the transformant AD-VaPAT1 + BD-VaIDD3 grew on the quadruple dropout medium (SD/Trp-Leu-His-Ade) supplemented with 200 ng/mL AbA and showed β-galactosidase activity (Figure 2, C). The interaction between VaPAT1 and VaIDD3 was further validated in vivo using bimolecular fluorescence complementation (BiFC) analysis in N. benthamiana. The results showed that a significant proportion of cells co-transformed with VaPAT1-nYFP (nYELLOW FLUORESCENT PROTEIN) + VaIDD3-cYFP and VaPAT1-cYFP + VaIDD3-nYFP displayed intense yellow fluorescence, indicative of an interaction between the tested protein combinations (Figure 2, D). Together, both results from Y2H and BiFC support the interaction between VaIDD3 and VaPAT1.
VaIDD3 have complementary roles with VaPAT1in the cold stress responses of grape
The interaction between VaIDD3 and VaPAT1 also prompted us to investigate the role of VaIDD3 in grapevine cold stress tolerance. First, VaIDD3 was overexpressed in grapes, the positive calli were selected by kanamycin (Supplemental Figure S4). RT-qPCR analysis showed that VaIDD3 expression was up-regulated in the transgenic calli line, with a consistent FC increase of approximately 100-fold when compared with the EV line. The calli were used for cold tolerance evaluation using several key physiological indices including LTE and relative electrolyte leakage (Supplemental Figure S4). Lower (−0.36°C), but not significant LTE was observed in OE-VaIDD3 lines (−7.06°C) when compared with the EV (−6.70°C) control (Supplemental Figure S4). Conversely, OE-VaIDD3 lines showed significantly lower electrolyte leakage compared with EV transgenic lines at cold (4°C, 4 h) temperatures. These results indicated that VaIDD3 may have complementary roles in the cold stress responses of grape with VaPAT1.
Modulation of cold-responsive gene expression in VaPAT1 and VaIDD3 -overexpressing grape
To gain insights into VaPAT1-mediated networks, RNA-seq of OE-VaPAT1 and control EV calli under normal temperature was performed. Differential expression (DE) analysis revealed a total of 2,494 up- and 1,406 down-regulated genes (FDR < 0.05, | log2FC | > 1) in OE-VaPAT1 lines compared with the control EV calli (Supplemental Table S3). MapMan functional enrichment analysis of significantly up-regulated genes revealed that terms related to abiotic stress (BIN20.2, FDR < 2.64E−15), raffinose metabolism (BIN3.1, FDR < 5.70E−06), glycolysis (BIN4, FDR < 1.96E−05), transcriptional regulation (BIN27.3, FDR < 3.43E−05), and hormone metabolism (BIN17, FDR < 1.33E−04) were among the most highly enriched.
To test if the transcriptome changes in OE-VaPAT1 lines relate to cold stress, RNA-seq of normal and cold-treated EV calli (4EV) was performed. DE analysis identified 2,307 up- and 1,687 down-regulated genes (FDR < 0.05, | log2FC | > 1) in 4EV compared with the control (Supplemental Table S4). Interestingly, many highly enriched terms in both up- and down-regulated genes lists were identical to those found in OE-VaPAT1 such as abiotic stress (BIN20.2, FDR < 1.38E−11), raffinose metabolism (BIN3.1, FDR < 1.23E−04), and transcriptional regulation (BIN27.3, FDR < 7.52E−05).
The interaction between VaPAT1 and VaIDD3 also prompted us to investigate VaIDD3-mediated transcriptome modification. A total of 2,419 up- and 2,097 down-regulated DEGs were identified from OE-VaIDD3 lines compared against the control EV calli (Supplemental Table S5). Transcriptional regulation (BIN27.3, FDR < 4.37E−08), hormone metabolism (BIN17, FDR < 7.60E−05), and protein post-translational modification (BIN29.4, FDR < 2.67E−03) were among the most highly enriched in upregulated genes.
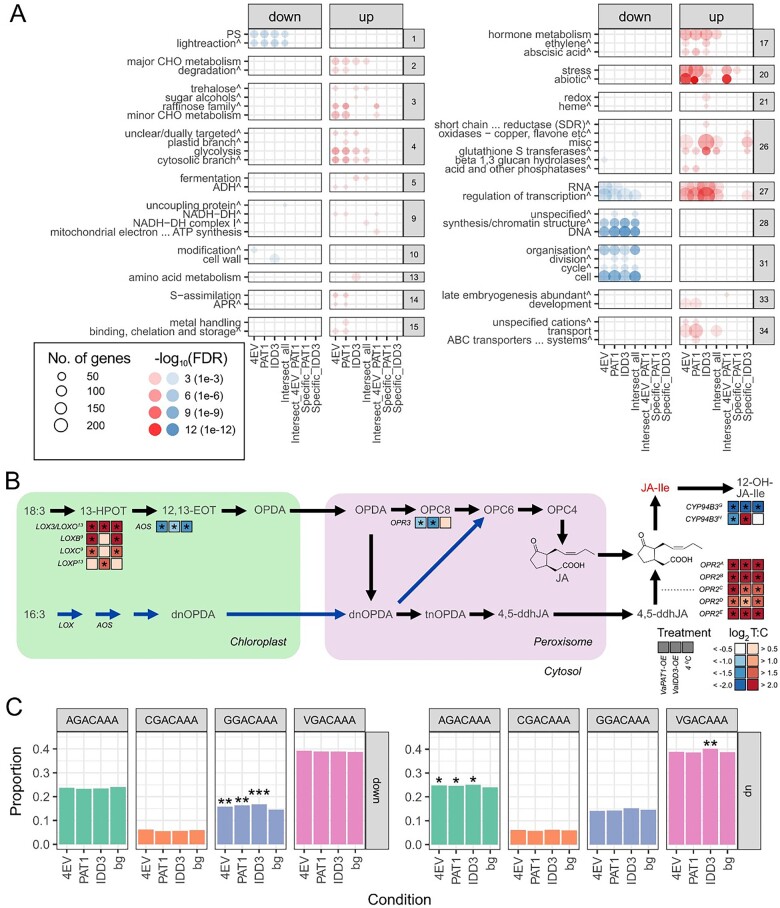
Comparative analysis of functional enrichment was performed to identify the common and unique pathways among OE-VaPAT1, 4EV, and OE-VaIDD3 lines (Figure 3, A andSupplemental Table S6). Enriched functional categories associated with hormone metabolism, especially JA synthesis including 12-oxo-phytodienoic acid (OPDA) reductase (BIN 17.7.1.5), sugar metabolism including raffinose synthases (BIN 3.1.2) and trehalose-6-phosphate synthase/phosphatase (BIN 3.2), posttranslational modification (BIN 24.1), and regulation of transcriptional activity by WRKY TFs (BIN 27.3.32) were identical in OE-VaPAT1, 4EV, and OE-VaIDD3 lines. Nonetheless, several enriched BIN categories were unique in OE-VaPAT1 and OE-VaIDD3 lines. For example, glutathione S-transferases (BIN 26.9) and regulation of transcriptional activity by Homeobox TFs (BIN 27.3.22) were enriched unique in OE-VaIDD3 lines.
Figure 3.
Comparative transcriptome analysis of grapevine Cold (4EV), OE-VaPAT1 (PAT1), and OE-VaIDD3 (IDD3) DE genes. A, Enriched BIN categories (FDR < 0.05, BIN depth ≤ 1). The circle size and the circle color depict the number of genes in each enriched category and the parent BIN category of each BIN, respectively. Enrichment scores (expressed as—log10 FDR) for each BIN category are shown. Red and blue colors indicate upregulated and downregulated genes, respectively. B, Simplified scheme of two alternative plant JA biosynthesis pathways. Only differentially expressed pathway genes are indicated. See main text for gene accessions. Pathway constituents (indicated in gray text) are 18:3, α-linolenic acid; 16:3, hexadecatrienoic acid; 13-HPOT, (13S)-hydroperoxyoctadecatrienoic acid; 12,13-EOT, 12,13(S)-epoxy-octadecatrienoic acid; OPC-6, 6-(3-oxo-2-(pent-2-enyl)cyclopentyl)hexanoic acid; OPC-4, 4-(3-oxo-2-(pent-2-enyl)cyclopentyl)butanoic acid; dnOPDA, dinor-OPDA; tnOPDA, tetranor-OPDA; 4,5-ddhJA, 4,5-didehydro-JA; JA-Ile, JA-Isoleucine; and 12-OH-JA-Ile, 12-Hydroxy-JA-Ile. The bioactive JA (JA-Ile) is indicated in red text. Black and blue pathway arrows define the octadecanoid and parallel hexadecanoid pathway, respectively. Asterisks denote significant DE (absolute log2 FC > 1, FDR < 0.05) between contrast. LOX, lipoxygenase; OPR, OPDA reductase; CYP94B3, JA-Ile-12-hydroxylase. C, Proportion of DEGs containing the IDD-box (VGACAAA, V = A/C/G) CRE in the promoter region of 4EV, OE-VaPAT1, OE-VaIDD3 DE genes. *FDR < 5.0E−02, **FDR between 1.0E−02 and 1.0E−03, and ***FDR < 4.0E−04 indicate significantly enriched CRE.
Further interrogation of the jasmonic acid (JA) biosynthetic pathway also revealed several DE genes involved in its biosynthesis (i.e. LOX, lipoxygenase; OPR, OPDA reductase; AOS, allene oxide synthase) and degradation (i.e. CYP94B3; Figure 3, B andSupplementary Tables S4, S5). For example, of the 18 predicted grapevine lipoxygenase (LOX) genes (Podolyan et al., 2010), four were upregulated (e.g. LOX3/VvLOXO, VIT_209s0002g01080; VvLOXP, VIT_201s0010g02750; VvLOXB, VIT_214s0128g00790; VvLOXC, VIT_214s0128g00780) while none were downregulated in either the cold, OE-VaPAT1, or OE-VaIDD3 lines (Figure 3, B). Interestingly, LOX3/VvLOXO, the gene encoding a functional 13-LOX (Podolyan et al., 2010), was highly upregulated in all three treatments compared with the control EV calli. Conversely, of the six predicted grapevine AOS, one (VIT_218s0001g11630) was significantly downregulated in three treatments compared with the control EV calli. The grapevine genome possesses four, seven, and one predicted 12-oxophytodienoate reductase 1 (OPR1), OPR2, and OPR3 genes, respectively. Interestingly, the gene encoding OPR3 (VIT_211s0016g01230) involved in the reduction of OPDA to 8-(3-oxo-2-(pent-2-enyl)cyclopentyl)octanoic acid (OPC-8) in the peroxisome (Chini et al., 2018) was downregulated in OE-VaPAT1 and OE-VaIDD3 lines compared with the control EV calli. In contrast, those implicated in the alternative JA biosynthetic pathway involving the reduction of 4,5-didehydro-JA to JA by OPR2 after release into the cytosol (e.g. named OPR2A-E for simplicity – OPR2A, VIT_218s0041g02020; OPR2B, VIT_218s0041g02060; OPR2C, VIT_218s0041g02080; OPR2D, VIT_218s0041g02270; and OPR2E, VIT_218s0122g01170) were significantly upregulated in all three treatments. We did not detect any significant DE of known grapevine JA-conjugating GH3 enzymes (i.e. VvGH3-7 and VvGH3-9) involved in bioactive JA (JA-Ile) production (Böttcher et al., 2015). However, the gene encoding jasmonoyl-isoleucine-12-hydroxylase (CYP94B3) that catalyzes the formation of 12-Hydroxy-JA-Ile (12-OH-JA-Ile) from the bioactive JA-Ile, hence the attenuation of JA responses (Koo et al., 2011), was differentially modulated (Figure 3, B). The grapevine genome contains two predicted CYP94B3 enzymes (i.e. CYP94B3G, VIT_217s0000g01490; CYP94B3H and VIT_214s0068g02120) of which the former was highly downregulated in all three treatments compared with the control EV calli while the latter was differentially modulated in OE-VaPAT1 and OE-VaIDD3 lines.
As mentioned above, VaPAT1 may interact with VaIDD3 to regulate the expression of downstream genes (Feurtado et al., 2011; Fukazawa et al., 2014; Yoshida et al., 2014; Hirano et al., 2017; Hakoshima, 2018). The VGACAAA motif (V = A/C/G, named IDD-box here) was previously shown to be the binding sequence of IDD family members in Arabidopsis and several other plants (Kozaki et al., 2004; Colasanti et al., 2006; Seo et al., 2011; O’Malley et al., 2016; Kumar et al., 2019). Therefore, the promoters of DEGs in OE-VaPAT1, OE-VaIDD3, and 4EV (Cold) were analyzed to identify potential IDD binding motifs. Between the four motifs, AGACAAA was significantly enriched (P < 0.05) in the promoters of OE-VaPAT1, OE-VaIDD3, and 4EV up-regulated genes (Figure 3, C andSupplemental Table S7) while the GGACAAA motif was significantly enriched in the promoters of VaPAT1-OE (P < 0.01), OE-VaIDD3 (P < 0.001), and 4EV (P < 0.01) down-regulated DEGs (Figure 3, C). These results indicate some degree of overlap in the cold stress-related transcriptional regulation of VaPAT1 and VaIDD3 in grapevines.
Integrated gene co-expression network analysis of PAT1 in grapevine
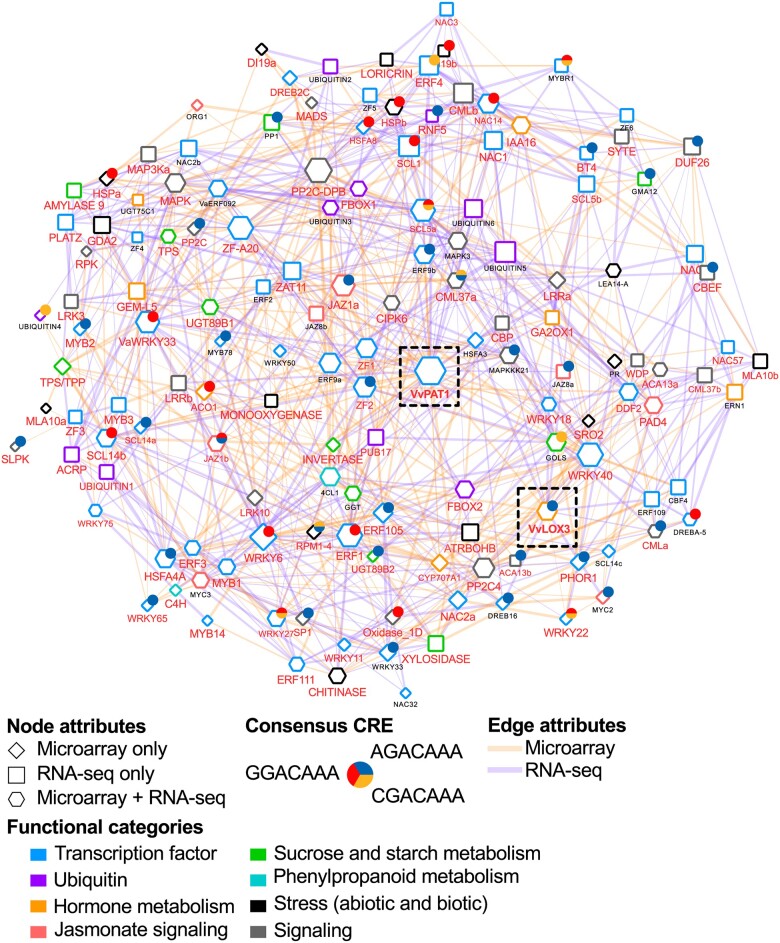
To gain insights into the transcriptional regulatory network of grapevine PAT1 and its role in cold stress response, we interrogated the latest microarray and RNA-seq datasets using PAT1 as “guide” genes. The top 300 most highly correlated genes (ranked by MR) were analyzed. Several relevant functional categories were identified along with the presence of IDD-box in the promoters of VaPAT1 co-expressed genes (Figure 4 and Supplemental Table S8). The genes related to hormone metabolism such as LOX3 were highly co-expressed. Sugar metabolism genes such as GALACTINOL SYNTHASE (GOLS); TREHALOSE-6-PHOSPHATE SYNTHASE, TPS; ALKALINE/NEUTRAL INVERTASE C, A/N-INVC (FYI) and AMYLASE 9 and those encoding various TF families such as AP2/ERFs (e.g. ERF1, ABR1, and ERF092), C2H2 (e.g. ZF1, ZAT11), GRAS (e.g. SCL1, SCL14a/b/c, SCL21), NAC (e.g. NTL9, ATAF1), and WRKY (e.g. WRKY22, WRKY33, WRKY40) were also identified. In addition, several signaling-related genes (e.g. CALMODULIN LIKE, CML37a; CBL-interacting protein kinase, CIPK6; MITOGEN-ACTIVATED PROTEIN KINASE, MPK3a) and Ubiquitin proteins (e.g. FBOX2, RNF5) are also of interest.
Figure 4.
Integrated gene co-expression subnetwork of grape PAT1. Node border color depicts common MapMan functional categories of co-expressed genes. Pie charts alongside the grapevine genes (right) represent the presence of IDD-box (VGACAAA, V = A/C/G) consensus CREs in the promoters of each corresponding gene. Blue, red, green, and node pie chart colors depict AGACAAA, GGACAAA, and CGACAAA CREs, respectively. The node shape depicts the co-expression relationship (light orange and blue edges) supported by both microarray and RNA-seq data (hexagon), microarray data only (diamond), and RNA-seq data only (square). Functional categories are represented by border color. Node size indicates connectivity. Gene labels in red depict consistent upregulation in at least one treatment (i.e. 4EV, OE-VaPAT1, or OE-VaIDD3 lines).
To ascertain whether the close relationship observed between PAT1 and its co-expressed genes is linked to a coordinated regulation of cold responses, we investigated their transcriptional responses to cold in our RNA-seq experiment (4EV) and from another comprehensive study of four grape varieties subjected to various cold treatment (accession numbers SAMN07617826-SAMN07617885; Londo et al., 2018). We observed that a large proportion of PAT1 co-expressed genes were upregulated in 4EV, OE-VaPAT1, and/or OE-VaIDD3 lines (Supplemental Table S8). The expression of a large proportion of PAT1 co-expressed genes (ca. 75%) was also induced in three treatments namely “acclimation” (4°C for 48 h), “acclimation freeze” (4°C for 48 h followed by “freeze”—3.0°C for 45 min), and “freeze” (−3.0°C for 45 min, no acclimation) across four cultivars of V. vinifera (Londo et al., 2018; Supplemental Figure S5 and Supplemental Table S9). These results suggest that PAT1 may be involved in the complex, yet coordinated regulation of cold responses. This may also relate to previous findings that it increases drought and salt tolerance in transgenic Arabidopsis (Yuan et al., 2016). Together, the RNA-seq and gene co-expression analysis provided strong candidates for the regulatory targets of VaPAT1.
VaPAT1 interacts with VaIDD3 to activate VaLOX3
Based on the co-expression network and RNA-Seq analysis, LOX3 was chosen as a plausible downstream target for the following reasons: in earlier studies, VvLOX3/VvLOXO encodes a functional 13-LOX enzyme involved in JA biosynthesis in grape (Podolyan et al., 2010) and implicated in various JA-mediated abiotic (e.g. wounding) and biotic (e.g. Botrytis and downy mildew infection) stress responses (Podolyan et al., 2010; Marchive et al., 2013). In this study, VvLOX3 showed strong co-expression with grapevine PAT1 in calli and leaves under cold treatment (Supplemental Figure S6), and its transcription was strongly induced by overexpression of both VaPAT1 and VaIDD3. A potential IDD binding site (AGACAAA) was also identified in its promoter. Thus, we hypothesized that VaPAT1 may involve in the transcriptional regulation of VaLOX3 during cold stress.
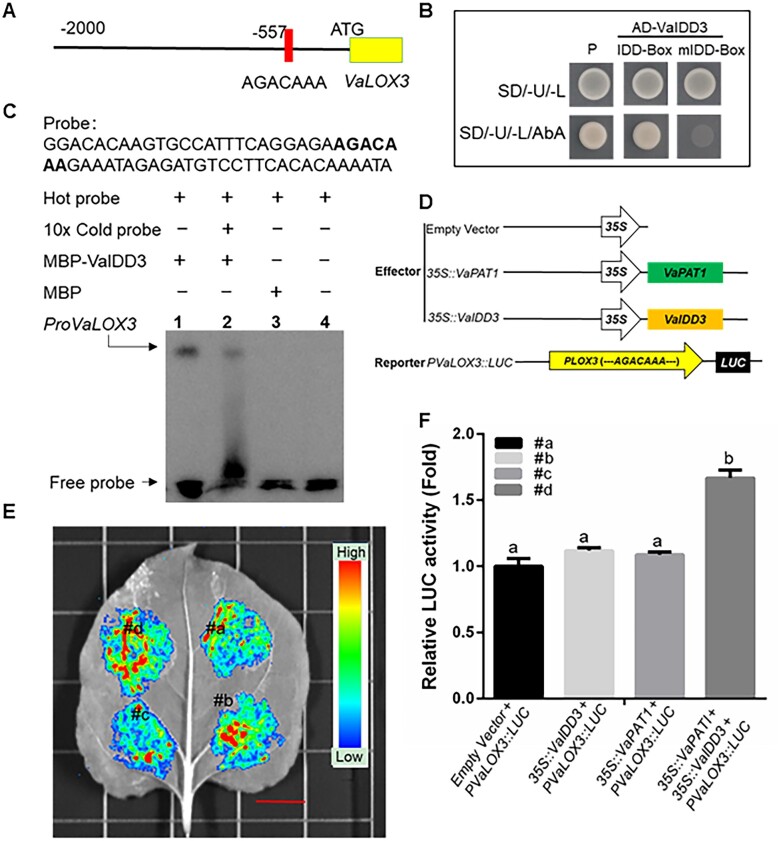
To test the hypothesis, the promoter (∼2,000 bp) of VaLOX3 from V. amurensis was cloned (The primer pairs are shown in Supplemental Table S1). A predicted IDD-box (AGACAAA) was found between positions −557 and −551 bp upstream of VaLOX3 ORF (Figure 5, A). Y1H assays showed that VaIDD3, but not VaPAT1, specifically binds to the IDD-box in the promoter sequence of VaLOX3 (Figure 5, B andSupplemental Figure S7, A). Subsequently, electrophoretic mobility shift assay (EMSA) was performed using VaLOX3 promoter (containing IDD-box) as the labeled probe. MBP-tagged VaIDD3 (MBP-VaIDD3) and GST-tagged VaPAT1 were purified and used for the DNA-binding assays. As shown in Figure 5, C, VaIDD3 bound to the IDD-box and the signal reduced when the unlabeled probe was added as a competitor (Figure 5, C, Lanes 1 and 2). Conversely, VaPAT1 failed to interact with the promoter containing IDD-box (Supplemental Figure S7, B). These results confirmed that VaIDD3 can bind to the VaLOX3 promoter in vitro.
Figure 5.
VaIDD3 bind to the IDD-box (AGACAAA) and VaPAT1/VaIDD3 complex promotes VaLOX3 transcription. A, Structure of the VaLOX3 promoter regions. IDD-box (AGACAAA) was identified at the −557 to −551 bp promoter position. B, Y1H analysis, using pGADT7-VaIDD3 as the prey, p3*IDDBox-AbAi, and p3*mIDDBox-AbAi as the baits and pGADT7-p53 and p53-AbAi as the positive controls. C, EMSA analysis of VaIDD3 binding to the VaLOX3 promoter. The 59-bp VaLOX3 promoter was labeled with biotin as the probe (Biotin-ProVaLOX3). The competition assay was performed using 10× unlabeled VaLOX3 promoter DNA (10× ProVaLOX3). The sequence of the biotin-labeled probe is shown and the IDD-box (AGACAAA) motif is highlighted in bold. D–F, LUC activity analysis, using 35S::VaPAT1 and 35S::VaIDD3 as the effector and pLOX3:LUC as reporters. The EV + PVaLOX3::LUC (#a) as the control. In (E), bar: 1 cm. The error bars indicate the SD from triplicate technical repeats. The letters above the bars indicate significant differences (P < 0.05, according to Duncan’s multiple range test).
To test if the interaction of VaPAT1 and VaIDD3 can activate the expression of VaLOX3, luciferase (LUC) reporter assay was performed in N. benthamiana leaves. Two constructs, 35S::VaPAT1 and 35S::VaIDD3 were used as effector and pLOX3:LUC as reporter. We observed that when either 35S::VaPAT1 or 35S::VaIDD3 were used, a relatively weak LUC activity was observed. However, co-transformation of VaPAT1 and VaIDD3 significantly enhanced the LUC activity by 1.7-fold compared with the control (P < 0.05, one-way ANOVA test; Figure 5, D–F). Collectively, the results indicate that VaPAT1 may interact with VaIDD3 to activate the downstream target gene, VaLOX3.
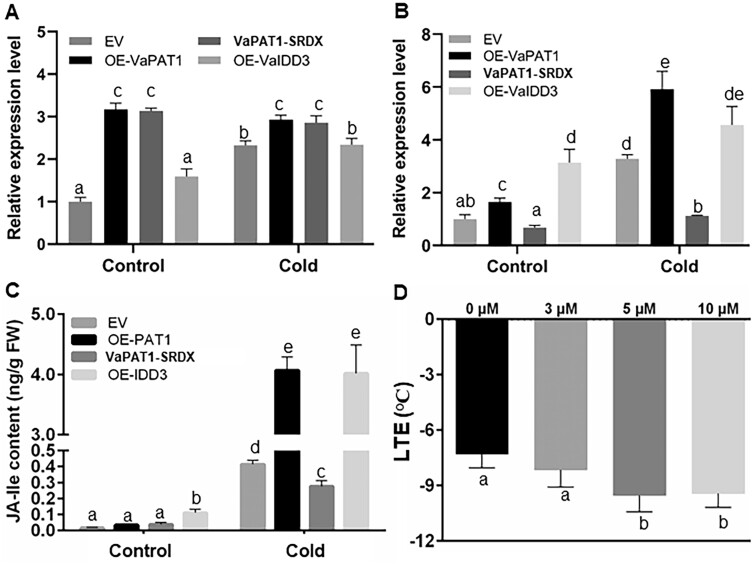
To ascertain whether VaPAT1 (and VaIDD3 co-operatively) modulate VaLOX3 expression in grape, we first investigated the expression of VaPAT1 and VvLOX3 in OE-VaPAT1, VaPAT1-SRDX, and OE-VaIDD3 lines. RT-qPCR analysis showed that VaPAT1 expression was significantly induced in OE-VaPAT1 and VaPAT1-SRDX lines compared with the EV in the control and cold conditions while no significant change was observed in OE-VaIDD3 lines (Figure 6, A). Conversely, the expression of VvLOX3 was significantly different across all lines but show consistent trends in the control and cold conditions. Both OE-VaPAT1 and OE-VaIDD3 lines showed elevated VvLOX3 expression while VaPAT1-SRDX showed significantly decreased levels (P < 0.05; Figure 6, B). Some of these findings (i.e. VaPAT1 and VaLOX3) also corroborated the expression observed in earlier RNA-seq analysis of 4EV, OE-VaPAT1, and OE-VaIDD3 experiments (Supplemental Tables S3–S5).
Figure 6.
Overexpression of VaPAT1 upregulates LOX3 expression and enhances JA accumulation during cold stress. A, Expression levels of PAT1 in transgenic lines. B, Expression levels of LOX3 during cold stress in transgenic lines. C, JA-Ile content in callus of EV, OE-VaPAT1, VaPAT1-SRDX, and OE-VaIDD3 lines under normal condition (Control) and cold-treated (Cold, 4°C, 4 h), respectively. D, LTEs of wild-type 41B callis grown in liquid GM medium or GM medium containing 3, 5, or 10 μM MeJA. The error bars indicate the SD from triplicate technical repeats. The letters above the bars indicate significant differences (P < 0.05, according to Duncan’s multiple range test).
VaPAT1 activates VaLOX3 expression and enhances JA-Ile accumulation in grape calli during cold stress and exogenous application of jasmonate enhances grape calli cold tolerance
Next, we determined whether the modulation of VvLOX3, a key gene in JA biosynthesis, contributes to an altered JAs (i.e. JA and bioactive JA-Ile) content in the different lines. Significant increase in JA-Ile was observed in EV control after the calli were subjected to cold treatment (4°C, 4 h; Figure 6, C and Supplemental Figure S8). No significant change in endogenous JA content was observed in all but one transgenic line (i.e. OE-VaIDD3) compared with the EV line when the calli were grown under normal growth conditions. Surprisingly, the JA-Ile content of OE-VaPAT1 and OE-VaIDD3 were significantly higher compared with the EV under cold treatment (4°C, 4 h; Figure 6, C). Accordingly, JA-Ile accumulation was significantly suppressed in VaPAT1-SRDX in accordance with the repression of VvLOX3 during cold stress (Figure 6, A–C). Similar dynamics to that of JA-Ile were observed for endogenous JA for OE-VaPAT1, VaPAT1-SRDX, and OE-VaIDD3 lines under normal growth conditions and cold treatment (4°C, 4 h; Supplemental Figure S8). To determine whether enhanced JA levels contribute to the cold tolerance of grape, LTEs of calli treated with exogenous MeJA were examined. We found that exogenous MeJA application at 5 and 10 µM significantly decrease LTE by 2.24°C and 2.13°C compared with the control calli (Figure 6, D). These findings highlight a close relationship between cold stress and endogenous JAs level in grape calli.
PAT1-ED mutants show decreased cold-tolerance, suppressed LOX3 expression, and JA content
To further ascertain the function of VaPAT1 in the transcriptional regulation of LOX3, we employed a robust CRISPR/Cas9 vector system for multiplex targeting of genomic sites in dicot plants (Ren et al., 2016; Osakabe et al., 2018). To achieve this, we first constructed two sites (sgRNA1 and 2) of PAT1 in the pYLCRISPR/Cas9 vector. Successful transformants were confirmed by detecting NPTII and Cas9 at the DNA and mRNA level (Supplemental Figure S9, A–C). To accurately verify the modification of DNA fragments, sequence analysis of EV-callis and sgRNA1/2-callis was performed. As the calli were chimeric, 20 clones of PCR amplicons from each sample were sequenced. The results showed that fragments from sgRNA1/2-calli contained indel mutations at the target site (Supplemental Figure S9, E and F). The examined sgRNA1/2-calli contained deletion or insertion mutations at target Site 2, with a high mutation rate of 50% (10 of 20 clones). Moreover, among the three kinds of mutation-containing sequences, one (33.33%, 1/3) exhibited stop codons on the 5′-region of their mRNA sequences (Supplemental Figure S9, G), which would result in premature translation termination. Furthermore, we detected a total of three variants, namely m1–m3 (Supplemental Figure S9), of which 90% consists of m1 and m2 variants.
To ascertain whether the observed PAT1-ED mutations are bonafide loss-of-function, two PAT1 variants (i.e. m1 and m2) were further characterized. As expected, both variants showed transcriptional activity in yeast as they retained the complete N-terminal domain (Supplemental Figure S10, A). This observation is consistent with earlier experiments demonstrating that the N-terminal of VaPAT1 has transcriptional activation activity (Figure 2, B). However, both variants were unable to (i) interact with VaIDD3 in Y2H experiments in yeast (Supplemental Figure S10, B) and (ii) activate VaLOX3 transcription in N. benthamiana (Supplemental Figure S10, C). Together, these results indicated that the two variants (i.e. m1 and m2) are loss-of-function for VaPAT1, the C-terminus of VaPAT1 was an effective target site, and frameshift mutations of VaPAT1 were likely to disrupt its gene expression and normal protein function.
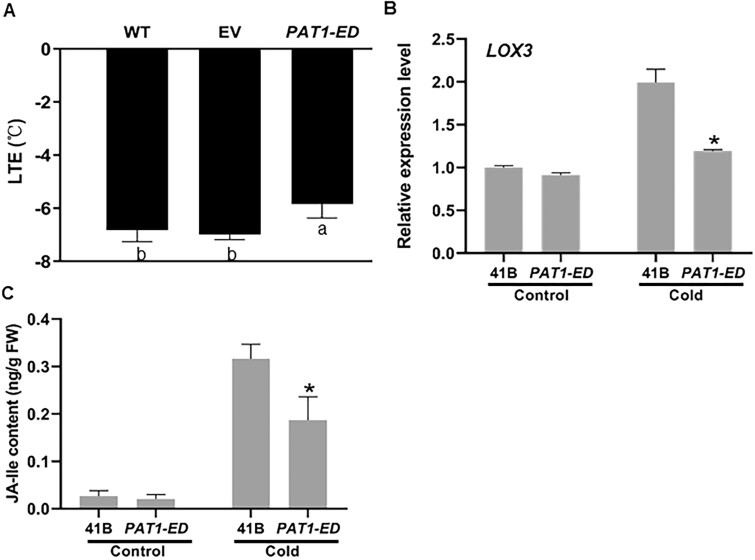
We hypothesized that disruption of PAT1 function may lead to altered LOX3 expression, thus affecting JA metabolism and cold tolerance. To test this, we used the line with truncated PAT1 lacking the C-terminal region (PAT1-ED; Supplemental Figure S9, G). PAT1-ED mutants exhibited significantly lower cold tolerance compared with the 41B/WT grape (−6.82°C) and EV control line (−6.98°C; Figure 7, A). The PAT1-ED mutants also exhibited decreased LOX3 expression especially under cold stress compared with 41B/WT grape (Figure 7, B). In agreement, the accumulation of JAs (i.e. JA and JA-Ile) during cold stress was also significantly decreased in PAT1-ED (Figure 7, C and Supplemental Figure S11). These results suggest that PAT1 mainly contributes to the induction of LOX3 during cold stress. Collectively, these results indicate that the involvement of VaPAT1 in cold tolerance may in part be due to its key role in the regulation of JA via VaLOX3 expression in grape. Nonetheless, a cooperative role of VaIDD3 in the regulation of JA and VaLOX3 expression with VaPAT1 is also indicated.
Figure 7.
The PAT1-ED revealed reduced expression of LOX3 and decreased cold tolerance. A, LTEs of 41B (WT), EV, and PAT1-ED (PAT1 mutants encoding truncated PAT1 lacking the C-terminal region, a loss-of-function mutation) lines. The error bars indicate the SD from three biological repeats and the letters above the bars indicate significant differences (P < 0.05, according to Duncan’s multiple range test). B, Expression levels of LOX3 in PAT1-ED line. The error bars indicate the SD from three biological repeats and asterisk indicates significant differences from the wild-type plants. *P < 0.01 (Student’s t test). C, JA-Ile content in callus of 41B/WT and PAT1-ED lines under normal condition (Control) and cold-treated (Cold, 4°C, 4 h), respectively. The error bars indicate the SD from three biological repeats and asterisk indicates significant differences from the wild-type plants. *P < 0.05 (Student’s t test).
Discussion
PAT1 has essential roles in grape during cold stress
GRAS TFs act as key regulatory hubs in adaptation to biotic and abiotic stresses, such as pathogens, cold, and hot (Fode et al., 2008; Navarro et al., 2008; Yuan et al., 2016; Yang et al., 2018). In this study, we observed a common theme of VaPAT1-mediated (over- and co-expression) networks genes involved in the transcriptional regulation (e.g. AP2/ERF, ZF, GRAS, HSF, NAC, and WRKY), sucrose, and starch metabolism (i.e. trehalose, raffinose, and starch), hormones (i.e. jasmonate, gibberelin, ethylene), and signaling (i.e. calcium and receptor-kinase related) in grapevine. Many of these genes are directly or indirectly implicated in the cold perception, signaling, low temperature-responsive transcriptional regulation, and regulation of metabolic response during cold stress/acclimation in land plants (Haake et al., 2002; Yamaguchi-Shinozaki and Shinozaki 2006; Knight and Knight 2012; Chai et al., 2019). They include well-known cold response pathways components (e.g. DREB2C, ZAT, CBF4), calmodulin, and calmodulin-like genes.
Meta-analysis of cold stress responses in grapevine further revealed that most co-expressed genes were induced during cold stress. Among the many co-expressed genes, we highlight the involvement of raffinose and starch metabolism, hormone metabolism, and the regulation of transcriptional processes. Previous studies have shown that these regulatory processes are important in the response to cold stress (Chinnusamy et al., 2007; Chai et al., 2019). We have recently shown that such approaches can help elucidate regulatory mechanisms exerted by grapevine AQUILO (Sun et al., 2018), VaERF092, and VaWRKY33 (Sun et al., 2019). These suggested that PAT1 may be involved in a complex regulatory network and play crucial roles in the regulation of grape cold stress response. These findings also point to its involvement in other abiotic stress. In this study, we focused on the downstream target, LOX3, however, our findings indicate many other high-confident targets that are worth validating in future studies.
VaPAT1/VaIDD3 complex synergistically regulate the expression of target genes
Although GRAS proteins function in transcriptional regulation, several GRAS proteins have been shown to play key roles in regulating by binding to IDD proteins (Fukazawa et al., 2014; Hirano et al., 2017; Hakoshima, 2018; Kumar et al., 2019). Hirano et al. (2017) found two GRAS complex (the SHR-SCR), which comprises an overall negatively charged surface potential, is unfavorable for binding to highly negatively charged DNA. Moreover, two reports suggested that the GRAS family proteins, SCL3 and DELLA, exclusively bind to IDDs (AtIDD3, AtIDD4, AtIDD5, AtIDD9, and AtIDD10) and play an antagonistic role in GA signaling (Fukazawa et al., 2014; Yoshida et al., 2014).
The GRAS domain comprises LHRI, VHIID, LHRII, PFYRE, and SAW modules. SHR possesses a relatively short N-terminal variable region, containing Gln, Thr, and Ser/His/Asn repeats (poly-Q, poly-T, and poly-S/H/N); the SCR possesses a longer N-terminal variable region, containing Ser/Pro, Ser, and Gln/Pro repeats (poly-S/P, poly-S, poly-Q/P), and a NLS (Hirano et al., 2017). The SHR-binding motif (R(K/R)DxxITHxAFCD) is widely conserved in the IDD family, and essential for SHR binding. It has been speculated that IDD family members could be further categorized into two groups, where one of the group comprises of SHR binding members (IDD1-13; Hirano et al., 2017). In grapes, VaPAT1-N (N-terminal) is similar to SHR and VaIDD3 comprise the SHR-binding motif (Supplemental Figure S3). We found that VaIDD3 and VaPAT1 (complete VaPAT1) can bind to each other in vivo, however, VaPAT1 lacking the C-terminal region, could not interact with VaIDD3. Like DELLA proteins (Yoshida et al., 2014), our findings highlight a key domain in the C-terminal of VaPAT1 required for proper protein interaction (Supplemental Figures S3, S10). Although we verified that VaPAT1 interacts with VaIDD3, we predict that other IDDs may interact with VaPAT1 to perform other regulatory functions.
Previous reports revealed the IDD genes encode putative proteins containing four zinc finger motifs (Colasanti et al., 2006; Xuan et al., 2013). The three ZFs, ZF1–ZF2–ZF3, of IDD proteins are important for DNA binding (Kozaki et al., 2004; Hirano et al., 2017). Most studies have shown that the ID1-cis motif (AGACAAA) represents the primary binding sequence of IDD family members (Kozaki et al., 2004; Colasanti et al., 2006; Kumar et al., 2019). Nonetheless, other variants exist. For example, AtIDD8 binding to GGACAAA sequence (Seo et al., 2011) and DNA affinity purification sequencing (DAP-seq) of several Arabidopsis IDDs highlighting the (A/C)GACAAA motif (O’Malley et al., 2016). Nonetheless, the AGACAA is often the main binding sequence of many different IDD family members (Kozaki et al., 2004; Yoshida et al., 2014). Consistent findings were also observed in this study. For example, only the AGACAAA motif was significantly enriched in upregulated DE genes of 4EV (cold), OE-VaPAT1, and OE-VaIDD3 lines (Figures 3, 4). Interestingly, the type of GGACAAA is significantly enriched in downregulated DE genes from 4EV (cold), OE-VaPAT1, and OE-VaIDD3, presumably because the GGACAAA motif was implicated in growth and development (Seo et al., 2011), however, further research would be needed to ascertain this functional link.
VaPAT1 regulates expression of the LOX3 related to JA biosynthesis in cold stress
JA is involved in cold stress responses and application of exogenous JA has been shown to improve cold tolerance and reduce cold injury in several economically important fruit crops, such as banana (Ba et al., 2016), mango (González-Aguilar et al., 2000), peach (Lei et al., 2003), and lemon (Siboza et al., 2014). Our results demonstrate that JA has a positive role in grape cold tolerance. The regulatory mechanisms of JA in the cold stress response have been investigated in several studies. For instance, Hu et al. (2013) showed that jasmonate acts as an upstream signal of the ICE-CBF/DREB1 transcriptional pathway to positively regulate freezing stress responses of Arabidopsis. However, one recent study challenged the role of ICE1 as an indispensable master regulator of the CBF/DREB1 genes (Thomashow and Torii 2020). This finding suggests that the factors responsible for the activation of jasmonate synthesis during cold stress remain unclear and more complex than previously thought. Indeed, some results suggest that many other TFs contribute to freezing tolerance that are independent of the CBF/DREB1 pathway (Park et al., 2015). Direct evidence for this are also plentiful. For instance, overexpression of ZAT12 and HSFC1 (Park et al., 2015) has been shown to increase plant freezing tolerance and upregulate the expression of many COR genes, including some that are coregulated by CBF/DREB1. Concordantly, our RNA-seq analysis also show that these genes (e.g. COR, HSF, and ZAT) were often coordinately up-regulated in OE-VaPAT1 and OE-VaIDD3 lines as well as cold treatment in comparison to the control EV calli under normal temperature (Figure 3, A andSupplemental Tables S3–S5).
Under low-temperature conditions, the expressions of key genes for JA synthesis such as OsLOX2 in rice and SlLOXD in tomato are induced (Du et al., 2013; Wang et al., 2016; Wasternack and Feussner 2018). However, the factors that are involved in the early response to cold stress to activate LOX3 for JA accumulation in grapes need to be further explored. Earlier studies have shown that grapevine VvLOX3 (VvLOXO) encodes a functional 13-LOX enzyme involved in JA biosynthesis (Podolyan et al., 2010) with wide implications in various JA-mediated abiotic and biotic stress responses such as wounding and various plant–pathogen interaction (Podolyan et al., 2010; Marchive et al., 2013). Marchive et al. (2013) demonstrated that grape transgenic lines overexpressing VvWRKY1 (VIT_217s0000g01280) were capable of modulating various JA biosynthetic and signaling pathway genes. Additional experimentation revealed that VvWRKY1 transactivated promoters of VvLOX3 and VvJAZ1, two key JA biosynthetic and signaling genes, respectively, and VvWRKY1 overexpression conferred a lower susceptibility to downy mildew infection. The present study identified VaPAT1 TF interaction with VaIDD3 and exhibited a positive effect on the reporter activity driven by the VaLOX3 promoter in vivo and in vitro (Figure 5). In cold-treated Amur grape apices, we also observed the induction of LOX3, in agreement with the temporal up-regulation of VaPAT1 (Figure 2 and Supplemental Figure S6). These results suggest that VaPAT1 is a transcriptional activator involved in the induction of VaLOX3 to increase JA accumulation in response to cold stress. A new role of LOX3 in grapevine abiotic stress response is also indicated.
In an earlier study, the expression of PAT1 in Amur grape was shown to be responsive to exogenous GA hormone and abiotic stress (i.e. cold, water deficit, and high salinity; Yuan et al., 2016). The latter findings prompted us to test whether VaPAT1 is also responsive to MeJA. We observed that expression of VaPAT1 rapidly increased after MeJA treatment (Supplemental Figure S12). This finding indicates a feedback regulation between the expression of PAT1 and JA, and thereby providing new insights into the cross-talk between GA and JA regulatory pathways.
In conclusion, low temperature can promote the biosynthesis of JA and endogenous JA can enhance the cold tolerance of grapevine. We isolated a stress-responsive GRAS gene (VaPAT1), which is located upstream of the JA signal pathway and is implicated as a positive regulator of cold tolerance. VaPAT1 overexpression enhanced the cold tolerance of grape calli. This finding indicated that VaPAT1, when induced by cold, may function as a key regulator that improves the cold tolerance of grapevine. Nonetheless, a role of VaIDD3 in grape cold tolerance is also indicated. Our study revealed that VaPAT1 regulates VaLOX3 by forming a complex with VaIDD3, and binding to the IDD-box (AGACAAA) in VaLOX3 promoter, to increase JA accumulation, leading to enhanced cold tolerance. These findings provide a model of how plants respond to cold stress before JA accumulation and transcriptional activation of downstream stress-inducible pathways in grape (Figure 8).
Figure 8.
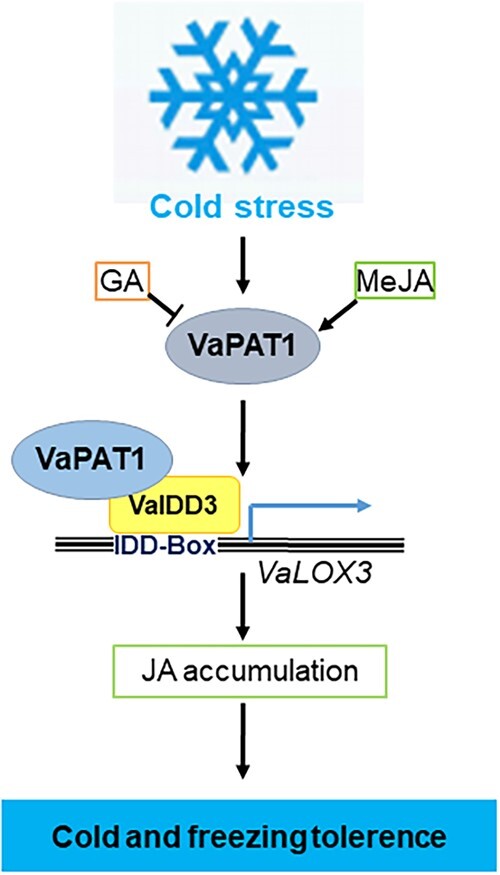
Model of VaPAT1 TF action during grape cold stress identified in this work. The model indicates that the cold stress-induced increase of VaPAT1 upregulates VaLOX3 expression (by forming a complex with IDD3) and increases JA accumulation, resulting in the transcriptional activation of stress-inducible genes and enhanced cold tolerance of grapevine. MeJA, methyl jasmonate.
Materials and methods
Plant materials and treatments
Micropropagated Vitis. amurensis and V. vinifera “Muscat Hamburg” plantlets were grown on half-strength Murashige and Skoog (1/2 MS, pH 5.8) as previously described (Zhao et al., 2017). Six-week-old plantlets were subjected to cold stress or supplied with plant growth regulators. For treatments with plant growth regulators, plantlet leaves were sprayed with 100 μM MeJA (MeJA, Sigma), whereas distilled water was used as control. Samples were collected at 0, 0.5, 1, 2, 4, and 8 h. Three biological replicates were collected for each sample. After the collection, all samples were immediately frozen in liquid nitrogen and then stored at −70°C.
An embryogenic cell suspension culture was initiated as described previously (Coutos-Thevenot et al., 1992). 41B embryogenic calli were cultured in 100 mL flasks fitted with 30 mL of liquid GM medium (Milieu MS 1/2 Macro [Ref: M0232 duchefa] supplemented with 1 g L−1 N-Z-Amine A [Ref: C7290 sigma], 1 mg L−1 2-naphthoxyacetic acid [NOA], 18 g L−1 Maltose [Ref: M0811 duchefa], 4.6 g L−1 Glycerol [Ref: G1345 duchefa], pH 5.8), and shaken at 120 rpm at 25°C in the dark. All suspension cells were sub-cultured every 7 d. For jasmonate treatment, 3, 5, or 10 μM MeJA was added to the GM medium and sub-cultured every 7 d.
Cloning and sequence analysis
The total genomic DNA was isolated from samples by using Plant Genomic DNA kit (Cat: DP305-03, Tiangen, Beijing, China). Total RNA was extracted from collected samples with Column Plant RNAout 2.0 (Cat: DP441, Tiangen, Beijing, China). First cDNA strands were synthetized by HiScript II 1st Strand cDNA Synthesis Kit (Cat: R212-01, Vazyme, Jiangsu, China). The open-reading frames (ORFs) of PAT1 and IDD3 were amplified from V. amurensis and V. vinifera “Muscat Hamburg” leaf cDNA, using the PCR primers PAT1-ORF-F/PAT1-ORF-R and DD3-ORF-F/IDD3-ORF-R by I5 High-Fidelity DNA Polymerase (Cat: TP001, Tsingke, Beijing, China; Supplemental Table S1). Sequences were aligned using DNAMAN 7.0.
RT-qPCR analysis
The HiScript II Q RT SuperMix for qPCR (Cat: R223-01, Vazyme, Jiangsu, China) was used to synthetize cDNA. Each sample was represented by three biological replicates and three technical replicates. 2−ΔΔCt method was used to calculate relative expression levels (Livak and Schmittgen 2001). The primers used for RT-qPCR are listed in Supplemental Table S1.
Transformation of 41B callis and generation of transgenic calli
The full-length VaPAT1 (VaPAT1-ORF-F/VaPAT1-ORF-R), VaPAT1-SRDX (VaPAT1-ORF-F/VaPAT1-SRDX-ORF-R), or VaIDD3 (VaIDD3-ORF-F/VaIDD3-ORF-R) ORFs, containing EcoR I and Xho I restriction sites (Trelief™ SoSoo Cloning Kit, Tsingke, Beijing, China), were ligated into the pSAK277 vector, downstream of the CaMV 35S promoter, and the vector was transformed into 41B calli referring to the protocol outlined in Zhao et al. (2017). For the method of A. tumefaciens-mediated transformation were according to Ren et al. (2016). The presence and expression of the transgene was confirmed by PCR and RT-PCR (primers for VaPAT1, VaIDD3, and NPTII are listed in Supplemental Table S1). Three lines were selected for further analyses.
Yeast assays
Yeast one-hybrid (Y1H) assay was performed using the Matchmaker Gold Y1H Library Screening System (Clontech) to examine the ability of VaPAT1 to bind with IDD-box motifs in the VaLOX3 promoter. Briefly, synthesized DNA fragment harboring three tandem copies of the IDD-box and its mutated form (mIDD-box) was cloned into the pAbAi vector to construct the baits. The primers IDDbox-F/IDDbox-R or mIDDbox-F/mIDDbox-R are available at Supplementary Table S1. The full-length CDS of VaIDD3 and VaPAT1 were ligated into the EcoRI-XhoI sites of the pGADT7 vector (Clontech). The Y1H assay was performed according to the Yeast Protocols Handbook (Clontech) to generate the prey vector (pGAD-VaIDD3 and pGAD-VaPAT1), respectively. The bait vectors were transformed into the Y1HGold yeast (Saccharomyces cerevisiae) strain. After selecting the transformants on SD/-Ura plates and determining the minimal inhibitory concentration of aureobasidin A (AbA) for the bait strains, pGAD-VaIDD3 and pGAD-VaPAT1 were introduced into the Y1HGold yeast strain, respectively. Positively co-transformed cells were screened on SD/-Leu medium supplemented with 200 ng mL−1 AbA and cultured at 30°C for 3 d. The positive control (pGADT7-p53 + p53-AbAi) was processed in the same manner.
Y2H assays were conducted using the Matchmaker Two-Hybrid System (Clontech). The AD vector pGADT7, the DNA-BD vector pGBDT7, and the yeast strain Y2HGold were utilized. As the prey, pGAD-VaPAT1 was used. The full-length CDS of VaIDD3, the full-length CDS of VaPAT1 (aa: 1–583), VaPAT1-D1 (aa: 1–212), and VaPAT1-D2 (aa: 213–583) was cloned, respectively, into the vector pGBKT7 and used as bait. The amplification primers used are shown in Supplemental Table S1. Selection of transformants was done on SD/-Trp media, and the trans-activation activity of each protein was evaluated according to their status of growth and the activity of X-α-Gal (5-bromo-4-chloro-3-indoxyl-α-d-galactopyranoside). The prey and bait vectors were co-transformed into Y2HGold cells and the transformants were identified according to the manufacturer’s instructions.
BiFC analysis in N. benthamiana
To generate BiFC constructs, the CDS of VaPAT1 and VaIDD3 was PCR amplified and ligated into 35S-SPYNE and 35S-SPYCE, resulting in VaPAT1-cYFP or VaPAT1-nYFP and VaIDD3-nYFP or VaIDD3-cYFP, respectively. EVs of BiFC constructs were used as negative controls. Primers used for generating fusion constructs are listed in Supplementary Table S1. All the constructs were transformed into the A. tumefaciens EHA105 strain and used for BiFC experiments.
The Agrobacterium growth conditions were in accordance to Zhao et al. (2017) and Agrobacterium solutions were adjusted to OD600 = 0.8 and then equally mixed before agroinfiltration. In this experiment, an Agrobacterium strain carrying the viral suppressor p19 was used to suppress gene silencing in transformed N. benthamiana leaves (Guan et al., 2017). The N. benthamiana plants were used for Agroinfiltration as previously described (Sheludko et al., 2007). The infiltrated N. bethamiana leaves were imaged using a Leica TCS SP5 (Leica, Germany) confocal microscope (laser: Argon, visible, 30%; emission bandwidth: 500–600 nm/650–750 nm; scan actuator gain: 4.72) 3 d after agroinfiltration. The excitation spectrum for imaging YFP fusions were 514 nm. All microscopy analyses were performed at least in triplicate with independent samples.
Subcellular localization
The ORF of VaPAT1 (excluding its termination codon) was fused to the green fluorescent (GFP) gene downstream of the Cauliflower mosaic virus (CaMV) 35S promoter in the pCAMBIA2300-35S-GFP vector. Resulting 35S:VaPAT1-GFP and the control 35S:GFP constructs were genetically introduced into N. benthamiana. Subcellular localization was observed using a Leica TCS SP5 confocal laser-scanning microscope (laser: Argon, visible, 30%; emission bandwidth: 500–600/650–750 nm; scan actuator gain: 6.74).
LUC assay
To confirm the effect of the PAT1/IDD3 complex on the transcriptional regulation and the interaction between VaIDD3 and the IDD-Box in the VaLOX3 promoter, LUC reporter assays were performed in N. benthamiana leaves. The VaLOX3 promoter (∼2,000 bp upstream of ATG) was amplified and cloned into the pCAMBIA1302-LUC to generate the reporter construct, pPLOX3-LUC. The pSAK277-VaPAT1 and pSAK277-VaIDD3 constructs were used as an effector. The effector and reporter constructs were transformed into Agrobacterium tumefaciens strain EHA105 and co-infected into N. benthamiana leaves by agroinfiltration, as previously described (Sheludko et al., 2007). Agrobacteria were cultured overnight and resuspended in infiltration buffer (10 mM MgCl2 and 10 mM MES) to a final OD600 of approximately 0.5. Agrobacteria were incubated at 28°C for 2 h before infiltration. LUC activities were detected using LUC Reporter Assay kit (Promega, Madison, WI, USA). The EV + PVaLOX3::LUC as the control. At least, three biological replicates were examined for each sample. All microscopy analyses were performed at least in triplicate with independent samples.
RNA sequencing analysis
RNA sequencing was performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China). Data processing and data analysis were performed as previously described (Meilong et al., 2020). Briefly, Trimmomatic v0.36 was used to remove low-quality reads, adapter sequences, and reads with ambiguous bases (Bolger et al., 2014). Parameters for obtaining high-quality reads were used: LEADING: 3, TRALING: 3, SLIDINGWINDOW: 4:15, and MINLEN: 90. All clean reads were mapped to the V. vinifera reference genome (12×; Jaillon et al., 2007) using the TopHat (v2.1.1; Kim et al., 2013), allowing no more than 2 bp mismatch. Gene annotation, expression quantification, and DE analysis were performed using the software Cufflinks (v2.2.1; Trapnell et al., 2012). Genes deemed differentially expression between any contrast of interests when the corresponding false discovery rate (FDR) is < 0.05 and the estimated absolute log2 (FC) > 1. MapMan BIN functional categories (Lohse et al., 2013) were identified using Fisher’s exact test and FDR correction as previously described for grape (Wong et al., 2017). Significantly enriched BIN categories were defined as categories having an FDR of < 0.05.
Integrated gene co-expression network analyses in grapevine
Grapevine gene co-expression networks (GCNs) were constructed as previously described (Vannozzi et al., 2018; Wong et al., 2018). Briefly, two separate transcriptome compendia consisting of RMA-normalized microarray (29K NimbleGen Grape Whole-genome platform) and FPKM (log2)-normalized RNA-Seq data sets were used. All biological replicates were averaged in the microarray and RNA-seq expression matrix, resulting in 356 and 236 columns (experiments), respectively, representing numerous and diverse experimental conditions. In this study, gene-centric co-expression (GCN) clusters were created for the candidate gene (i.e. VvPAT1), from both mutual rank (MR; Takeshi and Kengo 2009) microarray and RNA-seq GCNs. The top 300 co-expressed genes (ranked by MR value) in each microarray and RNA-seq GCNs were further shortlisted and analyzed. Shortlisted co-expressed genes in grapevine GCNs were tested for MapMan BIN functional category enrichment as previously described (Wong et al., 2017). Significantly enriched BIN categories were defined as categories having an FDR of < 0.05. Network visualization was performed using Cytoscape v3.5 (Shannon et al., 2003).
Gene expression meta-analysis of cold stress and related datasets in grapevine
Expression data of grapevine genes co-expressed under normal and cold/low temperature treatments were retrieved from Londo et al. (2018). Visualization of gene expression responses to treatments (expressed as log2 FC of treatment/control) was performed in R (http://www.r-project.org).
Cis-regulatory element analysis of promoter regions of co-expressed grape genes
The presence of the well-known IDD cis-regulatory elements (CREs), namely the IDD-box (VGACAAA, V = A/C/G), was screened for in the promoters (1.0 kb upstream of the transcription start site, TSS, and along both + and – strands) of co-expressed genes for exact pattern match using PatMatch (Yan et al., 2005). The position of each CRE identified and the total number of occurrences were recorded and tested for enrichment (based on hypergeometric distribution) and corrected for multiple hypothesis testing using FDR as previously described (Wong et al., 2017).
Electrophoretic mobility shift assay
The cDNA sequences of VaIDD3 from V. amurensis were inserted into the pMal-C2X (http://www.addgene.org) vector and transformed into Rosetta (DE3) chemically competent cell (Zoman Biotechnology Company, ZC125-1). Culture growth was induced at 16°C for 16 h and the cells were then suspended in cell buffer (10 mM Tris–HCl 7.4, 30 mM NaCl, 1 mM PMSF). The E. coli suspensions were sonicated for 20 min (in an ice bath) and centrifuged (12,000 g, 1 h, 4°C) and the supernatants were collected and incubated with MBP Amylose resins (NEB Corporation, Fond Du Lac, WI, USA, E8021V) at 4°C for 1 h. After discarding the supernatant, the remaining beads were washed three times with 500 μL per wash (20 mM Tris–HCl pH 7.4, 1 mM EDTA, 200 mM NaCl, 1 mM DTT) and eluted by elution buffer (20 mM Tris–HCl pH 7.4, 1 mM EDTA, 200 mM NaCl, 1 mM DTT, and 10 mM maltose). The cDNA sequence of VaPAT1 from V. amurensis was inserted into the pGEX-4T-2 vector. PBS buffer and Glutathione Sepharose (GE Corporation, New York City, NY, USA, 17075601) were used. The eluted protein was detected by Coomassie brilliant blue staining and western blot. For EMSA, biotin end-labeled double-stranded DNA probes were prepared by annealing complementary oligonucleotides. The oligonucleotides were heated at 95°C for 5 min, then at 72°C for 20 min, and left to cool to room temperature before use. EMSA was performed using the LightShift Chemiluminescent EMSA Kit (cat. no. 20148, Thermo Scientific, Waltham, MA, USA).
Design of sgRNA and assembly of CRISPR/Cas9 construct
The verified sequence of PAT1 was input into the online tool targetDesign (http://skl.scau.edu.cn/targetdesign/) to find potential Cas9 target sites and the output target sites were selected for designing target sgRNAs based on their location in the gene and off-target possibilities. The 20-bp sgRNA sequence was ligated into pYLCRISPR/Cas9 vector.
Cold tolerance evaluation of overexpressing grape calli
A system for differential thermal analysis (DTA) was used to assess the cold tolerance of Amur and 41B grape calli, using the protocol described by Sun et al. (2018). LTEs were obtained and used to evaluate cold tolerance.
Hormone detection
Fresh grape calli (41B) were harvested, weighted, immediately frozen in liquid nitrogen, and stored at −80°C until needed, respectively. Plant materials (50 mg fresh weight) were frozen in liquid nitrogen, ground into powder, and extracted with 0.5 mL methanol/water/formic acid (15:4:1, V/V/V) at 4°C with 2H5-JA (isoReag) added as an internal standard. The extracts were vortexed (10 min) and centrifuged at 14,000 rpm under 4°C for 5 min. The combined extracts were evaporated to dryness under nitrogen gas stream, reconstituted in 80% methanol (V/V), and filtrated (PTFE, 0.22 μm; Anpel, Shanghai, China) before LC–MS/MS analysis. The analytical conditions were as follows: HPLC: column, Waters ACQUITY UPLC HSS T3 C18 (1.8 µm, 2.1 mm * 100 mm); solvent system, water (0.04% V/V acetic acid): acetonitrile (0.04% V/V acetic acid); gradient program, 90:10 V/V at 0 min, 40:60 V/V at 5 min, 40:60 V/V at 7 min, 90:10 V/V at 7 min, 90:10 V/V at 10 min; flow rate, 0.35 mL/min; temperature, 40°C; injection volume: 2 μL. The effluent was alternatively connected to an ESI-triple quadrupole-linear ion trap (Q TRAP)-MS AB 6500+ QTRAP LC–MS/MS System, equipped with an ESI Turbo Ion-Spray interface, operating in both positive and negative ion modes and controlled by Analyst 1.6 software (AB Sciex, Framingham, MA, USA). The ESI source operation parameters were as follows: ion source, turbo spray; source temperature 500°C; ion spray voltage (IS) 5,500 V (Positive), −4,500 V (Negative); curtain gas (CUR) was set at 35.0 psi; the collision gas (CAD) was medium. A specific set of MRM transitions were monitored for each period according to the plant hormones eluted within this period. Identification of JAs (i.e. JA and JA-Ile) was performed with MetWare (http://www.metware.cn/) based on the AB Sciex QTRAP 6500 LC–MS/MS platform. Quantification of JAs was made using 2H5-JA as a reference for extraction recovery and standard curve determination purposes. Three replicates of each assay were performed (Cui et al., 2015).
Accession numbers
Sequence data from genes mentioned in this article can be found in the Vitis vinifera Genome (12×) annotation and Arabidopsis Genome or the GenBank data libraries under accession numbers: VvIDD3 (VIT_12s0028g03030), VvPAT1 (VIT_219s0014g04940), VvLOX3 (VIT_209s0002g01080), AtIDD3 (AT1G03840), MALATE DEHYDROGENASE (VvMDH, VIT_07s0005g03350), and VvACTIN (VIT_04s0044g00580).
Supplemental data
Supplemental Figure S1. Overexpression of VaPAT1 enhances cold tolerance in transgenic 41B grape calli.
Supplemental Figure S2. Subcellular localization of VaPAT1.
Supplemental Figure S3. Alignment of the VaIDD3 and VvIDD3 amino acid sequence with AtIDD3 proteins.
Supplemental Figure S4. Overexpression of VaIDD3 in 41B grape calli.
Supplemental Figure S5. The relationship between PAT1 and its co-expressed genes is linked to a coordinated regulation of cold responses.
Supplemental Figure S6. Expression pattern of VaLOX3 under cold stress in shoot apices and young leaves.
Supplemental Figure S7. Y1H assay and EMSA analysis of VaPAT1 binding to the VaLOX3 promoter.
Supplemental Figure S8. Overexpression of VaPAT1 upregulates LOX3 expression and enhances JA accumulation during cold stress.
Supplemental Figure S9. Identification of PAT1-ED positive transgenic callus and analysis of induced mutations in the target sequence.
Supplemental Figure S10. PAT1-ED mutations (m1 and m2) are loss-of-function.
Supplemental Figure S11 . JA content in callus of 41B/WT and PAT1-ED lines under normal (Control) and cold-treated (Cold, 4°C, 4 h) condition.
Supplemental Figure S12. Expression analyses of VaPAT1 under 100 μM MeJA treatments.
Supplemental Table S1. List of primer sequences used in this study.
Supplemental Table S2. Predicted IDD in grapevine.
Supplemental Table S3. Summary of transcriptomic data and DE results of OE-VaPAT1 compared with the EV at normal temperature.
Supplemental Table S4. Summary of transcriptomic data and DE results of the EV under cold conditions (4EV) compared with the EV at normal temperature.
Supplemental Table S5. Summary of transcriptomic data and DE results of OE-VaIDD3 compared with the EV at normal temperature.
Supplemental Table S6. Summary of enriched MapMan BIN categories of differentially expressed genes identified in (i) over-expression lines (e.g. OE-VaPAT1 and OE-VaIDD3) compared with the EV control under normal conditions and in (ii) the EV under cold stress compared to normal conditions.
Supplemental Table S7. Enrichment of IDD CRE (VGACAAA, V = A/C/G) in the promoter region of DE genes in 4EV, OE-VaPAT1, and OE-VaIDD3.
Supplemental Table S8. List and description of PAT1 co-expressed genes inferred from the microarray and RNA-seq datasets.
Supplemental Table S9. Meta-analysis of the transcriptional responses of VaPAT1 co-expressed genes to cold acclimation, acclimation freeze, and freeze treatments in leaves of four grapevine cultivars.
Supplementary Material
Acknowledgments
The authors thank Associate Prof. Sun Xiaoming (College of Horticulture, China Agriculture University) for providing the plasmid for the EMSA experiment.
Funding
This work was supported by the National Key Research and Development Program (2019YFD1002501) and the National Science Foundation of China (32025032).
Conflict of interest statement. The authors declare no competing interests.
Z.W. performed the experiments. D.C.J.W. and Y.W. performed the bioinformatics analysis. Z.W. and D.C.J.W. analyzed the data and wrote the paper. G.X., C.R., Y.L., and Y.K. established the transgenic system and organized the data. P.F., S.L., H.X., and Z.L. designed the research and supervised this study. All authors read and approved the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Zhenchang Liang (zl249@ibcas.ac.cn).
References
- Ba L, Jianfei K, Chen J, Lu W (2016) MaJAZ1 attenuates the MaLBD5-mediated transcriptional activation of jasmonate biosynthesis gene MaAOC2 in regulating cold tolerance of banana fruit. J Agric Food Chem 64:738–745 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218:683–692 [DOI] [PubMed] [Google Scholar]
- Böttcher C, Burbidge CA, di Rienzo V, Boss PK, Davies C (2015) Jasmonic acid-isoleucine formation in grapevine (Vitis vinifera L.) by two enzymes with distinct transcription profiles. J Integr Plant Biol 57:618–627 [DOI] [PubMed] [Google Scholar]
- Chai F, Liu W, Xiang Y, Meng X, Sun X, Cheng C, Liu G, Duan L, Xin H, Li S (2019) Comparative metabolic profiling of Vitis amurensis and Vitis vinifera during cold acclimation. Hortic Res 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12:444–451 [DOI] [PubMed] [Google Scholar]
- Chini A, Gimenez-Ibanez S, Goossens A, Solano R (2016) Redundancy and specificity in jasmonate signalling. Curr Opin Plant Biol 33:147–156 [DOI] [PubMed] [Google Scholar]
- Chini A, Monte I, Zamarreño AM, Hamberg M, Lassueur S, Reymond P, Weiss S, Stintzi A, Schaller A, Porzel A, et al. (2018) An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat Chem Biol 14:171–178 [DOI] [PubMed] [Google Scholar]
- Colasanti J, Tremblay R, Wong AY, Coneva V, Kozaki A, Mable BK (2006) The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutos-Thevenot P, Maes O, Jouenne T, Claude Mauro M, Boulay M, Deloire A, Guern J (1992) Extracellular protein patterns of grapevine cell suspensions in embryogenic and non-embryogenic situations. Plant Sci 86:137–145 [Google Scholar]
- Cui K, Lin Y, Zhou X, Li S, Liu H, Zeng F, Zhu F, Ouyang G, Zeng Z (2015) Comparison of sample pretreatment methods for the determination of multiple phytohormones in plant samples by liquid chromatography–electrospray ionization-tandem mass spectrometry. Microchem J 121:25–31 [Google Scholar]
- Du H, Liu H, Xiong L (2013) Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front Plant Sci 4:397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell A (2004) Freezing tolerance and injury in grapevines. J Crop Improv 10:201–235 [Google Scholar]
- Feurtado JA, Huang D, Wicki-Stordeur L, Hemstock LE, Potentier MS, Tsang EW, Cutler AJ (2011) The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 23:1772–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I, Wasternack C (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53:275–297 [DOI] [PubMed] [Google Scholar]
- Fode B, Siemsen T, Thurow C, Weigel R, Gatz C (2008) The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 20:3122–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Teramura H, Murakoshi S, Nasuno K, Nishida N, Ito T, Yoshida M, Kamiya Y, Yamaguchi S, Takahashi Y (2014) DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis. Plant Cell 26:2920–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Aguilar GA, Fortiz J, Cruz R, Baez R, Wang CY (2000) Methyl jasmonate reduces chilling injury and maintains postharvest quality of mango fruit. J Agric Food Chem 48:515–519 [DOI] [PubMed] [Google Scholar]
- Guan P, Ripoll JJ, Wang R, Vuong L, Bailey-Steinitz LJ, Ye D, Crawford NM (2017) Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc Natl Acad Sci U S A 114:2419–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130:639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakoshima T (2018) Structural basis of the specific interactions of GRAS family proteins. FEBS Lett 592:489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Nakagawa M, Suyama T, Murase K, Shirakawa M, Takayama S, Sun TP, Hakoshima T (2017) Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD. Nat Plants 3:17010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Lee LY, Xia K, Yan Y, Yu H (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19:884–894 [DOI] [PubMed] [Google Scholar]
- Howe GA (2018) Metabolic end run to jasmonate. Nat Chem Biol 14:109–110 [DOI] [PubMed] [Google Scholar]
- Hu Y, Jiang L, Wang F, Yu D (2013) Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25:2907–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Jiang Y, Han X, Wang H, Pan J, Yu D (2017) Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J Exp Bot 68:1361–1369 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight MR, Knight H (2012) Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol 195:737–751 [DOI] [PubMed] [Google Scholar]
- Kozaki A, Hake S, Colasanti J (2004) The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res 32:1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJK, Cooke TF, Howe GA (2011) Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-l-isoleucine. Proc Natl Acad Sci U S A 108:9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Le DT, Hwang S, Seo PJ, Kim HU (2019) Role of the INDETERMINATE DOMAIN Genes in Plants. Int J Mol Sci 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourieux F, Lecourieux D, Vignault C, Delrot S (2010) A sugar-inducible protein kinase, VvSK1, regulates hexose transport and sugar accumulation in grapevine cells. Plant Physiol 152:1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei F, Yonghua Z, Yanfen Z, Feng W, Lan Z, Zhaoxin L (2003) Methyl jasmonate reduces chilling injury and maintains postharvest quality in peaches. Agric Sci China 48:515–519 [DOI] [PubMed] [Google Scholar]
- Li J, Wang N, Xin H, Li S (2013) Overexpression of VaCBF4, a transcription factor from Vitis amurensis, improves cold tolerance accompanying increased resistance to drought and salinity in Arabidopsis. Plant Mol Biol Rep 31:1518–1528 [Google Scholar]
- Lindemose S, O’Shea C, Jensen MK, Skriver K (2013) Structure, function and networks of transcription factors involved in abiotic stress responses. Int J Mol Sci 14:5842–5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Huang W, Xian Z, Hu N, Lin D, Ren H, Chen J, Su D, Li Z (2017) Overexpression of SlGRAS40 in tomato enhances tolerance to abiotic stresses and influences auxin and gibberellin signaling. Front Plant Sci 8:1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Lohse M, Nagel A, Herter T, May P, Schroda M, Zrenner R, Tohge T, Fernie AR, Stitt M, Usadel BR (2013) Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ 37:1250–1258 [DOI] [PubMed] [Google Scholar]
- Londo JP, Kovaleski AP, Lillis JA (2018) Divergence in the transcriptional landscape between low temperature and freeze shock in cultivated grapevine (Vitis vinifera). Hortic Res 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C, Léon C, Kappel C, Coutos-Thévenot P, Corio-Costet MF, Delrot S, Lauvergeat V, Wang B, Tao J, Marchive C, et al. (2013) Over-expression of VvWRKY1 in grapevines induces expression of jasmonic acid pathway-related genes and confers higher tolerance to the downy mildew. PLoS ONE 8:e54185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilong X, Qian T, Yi W, Zemin W, Guangzhao X, Kirabi EG, Shaohua L, Zhenchang L (2020) Transcriptomic analysis of grapevine LEA gene family in response to osmotic and cold stress, and functional analyses of VamDHN3 gene. Plant Cell Physiol 261:775–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Matsui K, Ikeda M, Nakata M, Oshima Y, Nagatoshi Y, Ohme-Takagi M (2011) CRES-T, an effective gene silencing system utilizing chimeric repressors. Methods Mol Biol 754:87–105 [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lison P, Nemri A, Harberd NP, Jones JD (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18:650–655 [DOI] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165:1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Liang Z, Ren C, Nishitani C, Osakabe K, Wada M, Komori S, Malnoy M, Velasco R, Poli M, et al. (2018) CRISPR–Cas9-mediated genome editing in apple and grapevine. Nature Protoc 13:2844–2863 [DOI] [PubMed] [Google Scholar]
- Park S, Lee C-M, Doherty CJ, Gilmour SJ, Kim Y, Thomashow MF (2015) Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J 82:193–207 [DOI] [PubMed] [Google Scholar]
- Podolyan A, White J, Jordan B, Winefield C (2010) Identification of the lipoxygenase gene family from Vitis vinifera and biochemical characterisation of two 13-lipoxygenases expressed in grape berries of Sauvignon Blanc. Funct Plant Biol 37:767–784 [Google Scholar]
- Ren C, Liu X, Zhang Z, Wang Y, Duan W, Li S, Liang Z (2016) CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci Rep 6:32289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DE, Peng J, Harberd NPJB (2015) Plant GRAS and metazoan STATs: one family? Bioessays 22:573–577 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Ryu J, Kang SK, Park CM (2011) Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J 65:418–429 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheludko YV, Sindarovska YR, Gerasymenko IM, Bannikova MA, Kuchuk NV (2007) Comparison of several Nicotiana species as hosts for high-scale Agrobacterium-mediated transient expression. Biotechnol Bioeng 96:608–614 [DOI] [PubMed] [Google Scholar]
- Siboza XI, Bertling I, Odindo AO (2014) Salicylic acid and methyl jasmonate improve chilling tolerance in cold-stored lemon fruit (Citrus limon). J Plant Physiol 171:1722–1731 [DOI] [PubMed] [Google Scholar]
- Sun X, Jones WT, Rikkerink EH (2012) GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling. Biochem J 442:1–12 [DOI] [PubMed] [Google Scholar]
- Sun X, Matus JT, Wong DCJ, Wang Z, Chai F, Zhang L, Fang T, Zhao L, Wang Y, Han Y, et al. (2018) The GARP/MYB-related grape transcription factor AQUILO improves cold tolerance and promotes the accumulation of raffinose family oligosaccharides. J Exp Bot 69:1749–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Zhang L, Wong DCJ, Wang Y, Zhu Z, Xu G, Wang Q, Li S, Liang Z, Xin H (2019) The ethylene response factor VaERF092 from Amur grape regulates the transcription factor VaWRKY33, improving cold tolerance. Plant J 99:988–1002 [DOI] [PubMed] [Google Scholar]
- Sun X, Zhao T, Gan S, Ren X, Fang L, Karungo SK, Wang Y, Chen L, Li S, Xin H (2016) Ethylene positively regulates cold tolerance in grapevine by modulating the expression of ETHYLENE RESPONSE FACTOR 057. Sci Rep 6:24066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshi O, Kengo K (2009) Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res 16:249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF, Torii KU (2020) SCREAMing twist on the role of ICE1 in freezing tolerance. Plant Cell 32:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannozzi A, Wong DCJ, Holl J, Hmmam I, Matus JT, Bogs J, Ziegler T, Dry I, Barcaccia G, Lucchin M (2018) Combinatorial regulation of stilbene synthase genes by WRKY and MYB transcription factors in grapevine (Vitis vinifera L.). Plant Cell Physiol 59:1043–1059 [DOI] [PubMed] [Google Scholar]
- Wang F, Guo Z, Li H, Wang M, Onac E, Zhou J, Xia X, Shi K, Yu J, Zhou Y (2016) Phytochrome A and B function antagonistically to regulate cold tolerance via abscisic acid-dependent jasmonate signaling. Plant Physiol 170:459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111:1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Feussner I (2018) The oxylipin pathways: biochemistry and function. Annu Rev Plant Biol 69:363–386 [DOI] [PubMed] [Google Scholar]
- Wong DCJ, Lopez Gutierrez R, Gambetta GA, Castellarin SD (2017) Genome-wide analysis of cis-regulatory element structure and discovery of motif-driven gene co-expression networks in grapevine. DNA Res 24:311–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DCJ, Zhang L, Merlin I, Castellarin SD, Gambetta GA (2018) Structure and transcriptional regulation of the major intrinsic protein gene family in grapevine. BMC Genomics 19:248–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Zhu W, Wang L, Xiang Y, Fang L, Li J, Sun X, Wang N, Londo JP, Li S (2013) Genome wide transcriptional profile analysis of Vitis amurensis and Vitis vinifera in response to cold stress. PLoS ONE 8:e58740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14(Suppl):S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Jiao Y, Li R, Zhang N, Xiao D, Ding X, Wang Z (2014) Chinese wild-growing Vitis amurensis ICE1 and ICE2 encode MYC-type bHLH transcription activators that regulate cold tolerance in Arabidopsis. PLoS ONE 9:e102303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan YH, Priatama RA, Huang J, Je BI, Liu JM, Park SJ, Piao HL, Son DY, Lee JJ, Park SH, et al. (2013) Indeterminate domain 10 regulates ammonium-mediated gene expression in rice roots. New Phytol 197:791–804 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803 [DOI] [PubMed] [Google Scholar]
- Yan T, Yoo D, Berardini TZ, Mueller LA, Weems DC, Weng S, Cherry JM, Rhee SY (2005) PatMatch: a program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res 33:W262–W266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Gao X, Ma K, Li D, Jia C, Zhai M, Xu Z (2018) The walnut transcription factor JrGRAS2 contributes to high temperature stress tolerance involving in Dof transcriptional regulation and HSP protein expression. BMC Plant Biol 18:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Duan G, Li C, Liu L, Han G, Zhang Y, Wang C (2019) The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front Plant Sci 10:1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Hirano K, Sato T, Mitsuda N, Nomoto M, Maeo K, Koketsu E, Mitani R, Kawamura M, Ishiguro S, et al. (2014) DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc Natl Acad Sci U S A 111:7861–7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Fang L, Karungo SK, Zhang L, Gao Y, Li S, Xin H (2016) Overexpression of VaPAT1, a GRAS transcription factor from Vitis amurensis, confers abiotic stress tolerance in Arabidopsis. Plant Cell Rep 35:655–666 [DOI] [PubMed] [Google Scholar]
- Zhao T, Wang Z, Su L, Sun X, Cheng J, Zhang L, Karungo SK, Han Y, Li S, Xin H (2017) An efficient method for transgenic callus induction from Vitis amurensis petiole. PLoS ONE 12:e0179730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167:313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.