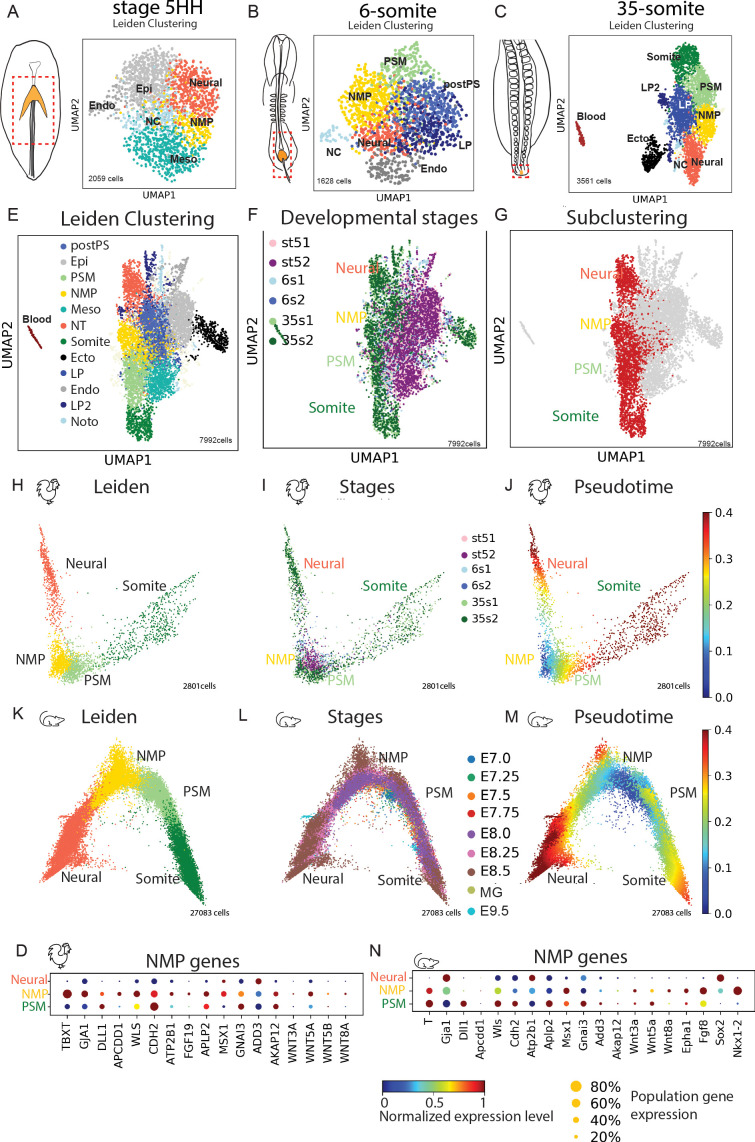
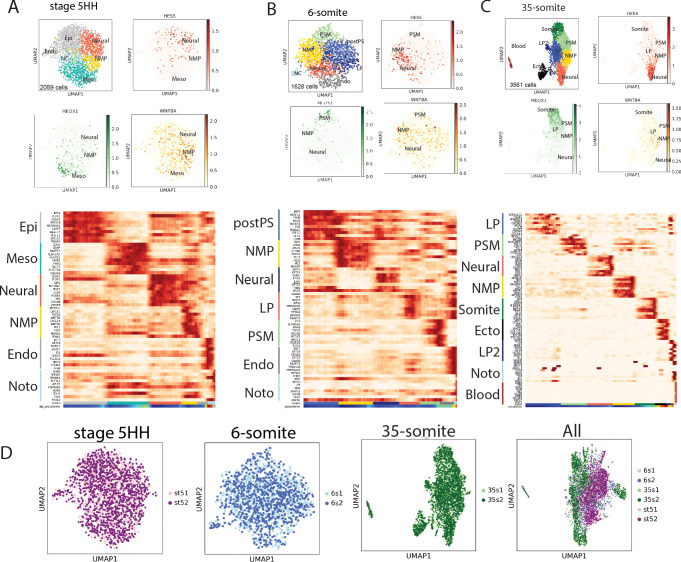
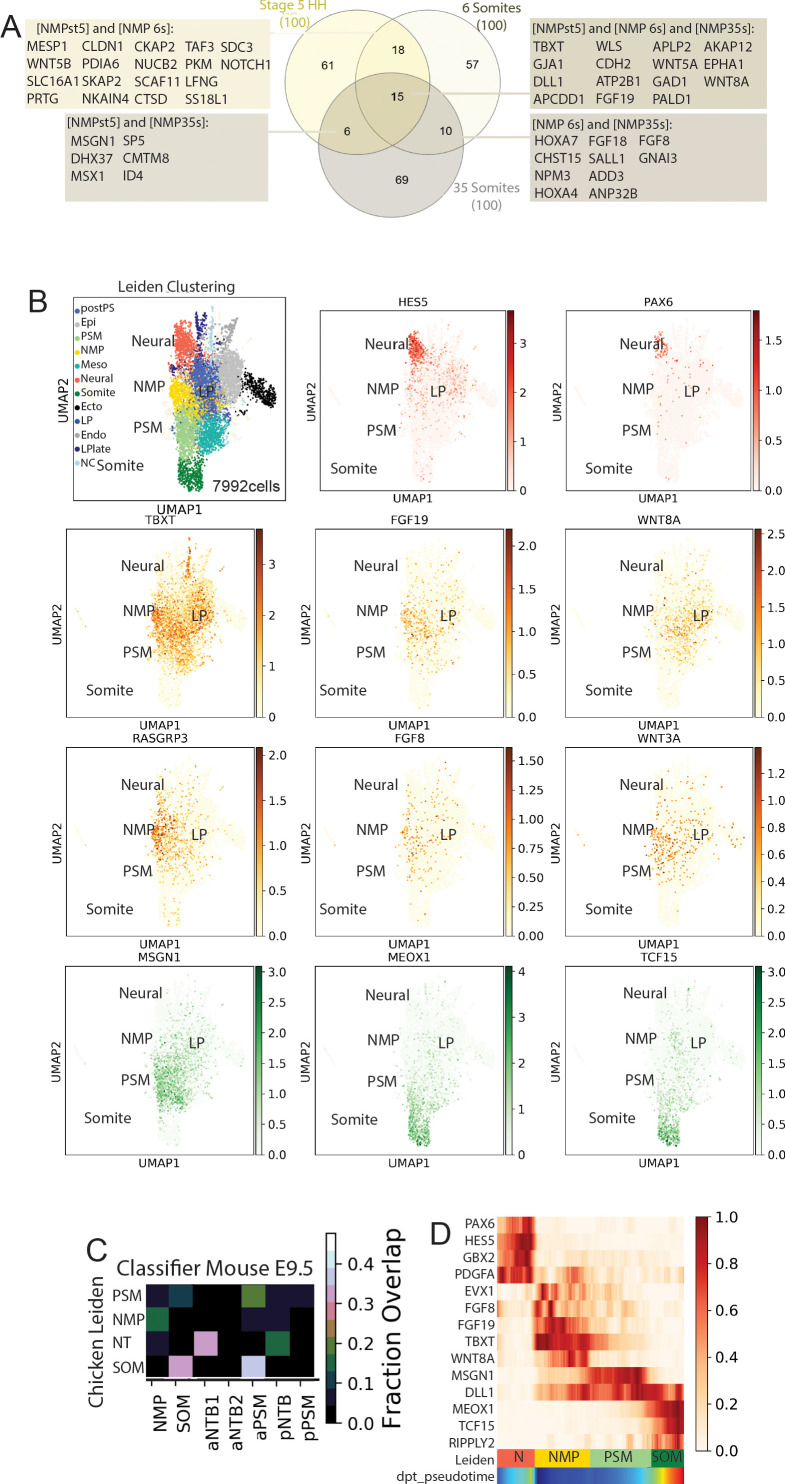
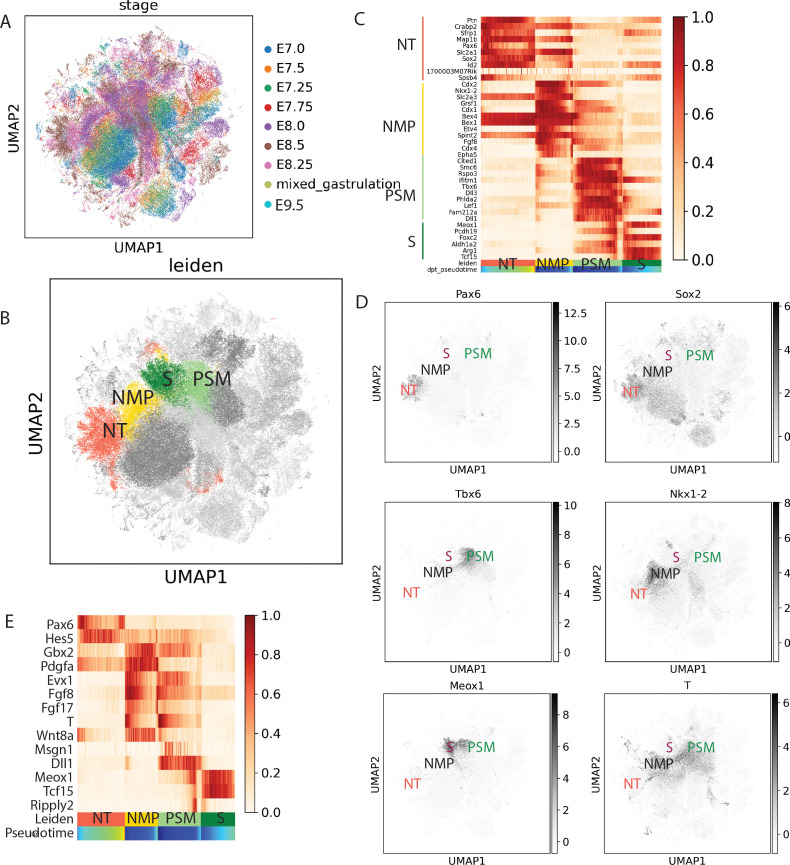
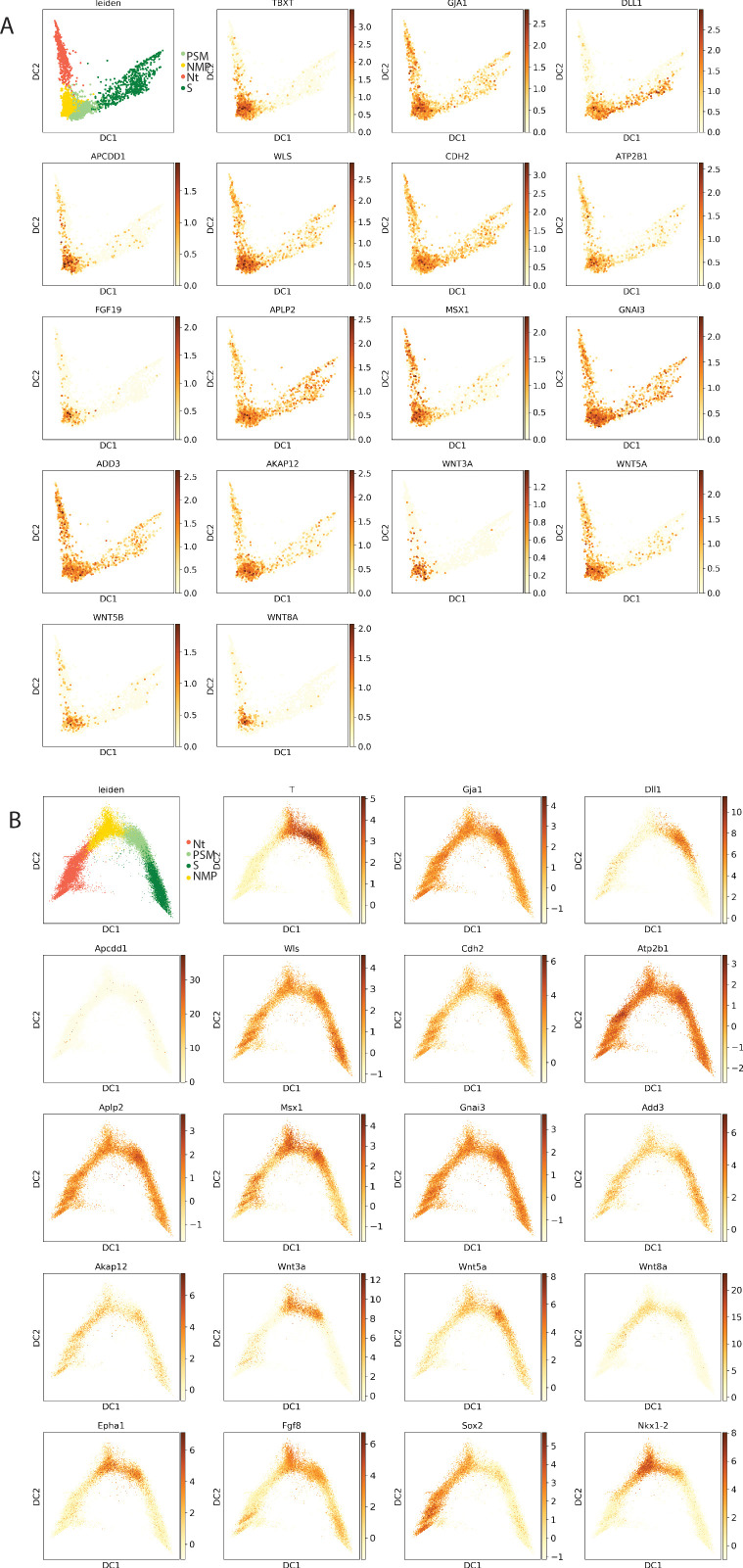
Figure 3. Single-cell RNA-sequencing (scRNAseq) analysis of the posterior tissue precursors during posterior axis formation.
(A–C) (left) Diagrams of a stage 5HH (A), a 6-somite (B), and a 35-somite (C) chicken embryo showing the region dissected and analyzed by scRNAseq in the hatched red boxes, which includes the NMP territory (in gold). (Right) k-NN graph showing the 2059 cells sequenced from stage 5HH embryos (A), the 1628 cells sequenced from 6-somite embryos (B), and the 3561 cells sequenced from 35-somite embryos (C) visualized with Uniform Manifold Approximation and Projection (UMAP). Single cells are colored based on Leiden clustering identities. (D) Dotplot showing the expression level of NMP signature genes in the chicken neural, NMP, and PSM clusters. (E–G) k-NN graphs combining all sequenced cells from stage 5HH, 6-somite and 35-somite chicken embryos (total: 7992 cells), visualized with UMAP and colored following Leiden clustering to show cell identities (E), or by developmental stage (F) or to show cells of the neural, NMP, PSM, and somite clusters (in red), which were used for the subsequent analysis shown in (H–J). Note that cells in the tan color belong to different cluster identities with less than 50 cells, and we decided to show them to represent the entirety of the data but do not analyze them in our study. (H–J) k-NN graphs showing cells of the chicken neural, NMP, PSM, and somite clusters extracted based on the analysis shown in (E–G) (total: 2801 cells) from stage 5HH, 6-somite, and 35-somite visualized with diffusion map (diffmap) and analyzed using Leiden clustering to show cell identities that include neural, NMP, PSM, and somite (H), as well as the developmental stage (I) or pseudo-temporal ordering with the NMP cluster as the starting node (J). (K–M) k-NN graphs showing cells of the mouse neural, NMP, and PSM clusters extracted based on the analysis shown in Figure 3—figure supplement 3 (total: 27,083 cells) from stage E7.0–E9.5 visualized with diffusion map (diffmap) and analyzed using Leiden clustering to show cell identities that include neural, NMP, and PSM (K), as well as the developmental stage (L) or pseudo-temporal ordering with the NMP cluster as the starting node (M). (N) Dotplot showing the expression level of NMP signature genes in mouse in the neural, NMP, and PSM clusters. Ecto: ectoderm; Endo: endoderm; Epi: epiblast; Meso: mesoderm; LP: lateral plate; NMP: neuromesodermal progenitors; NT: neural tube; NC: notochord; PSM: presomitic mesoderm; postPS: posterior PS; SOM: somite; LP2: lateral plate 2; MG: mixed gastrulation in (D, N). Circle size shows the percentage of cells expressing the gene in the cluster. Color shows the normalized level of expression. Normalization is done by gene across the clusters.