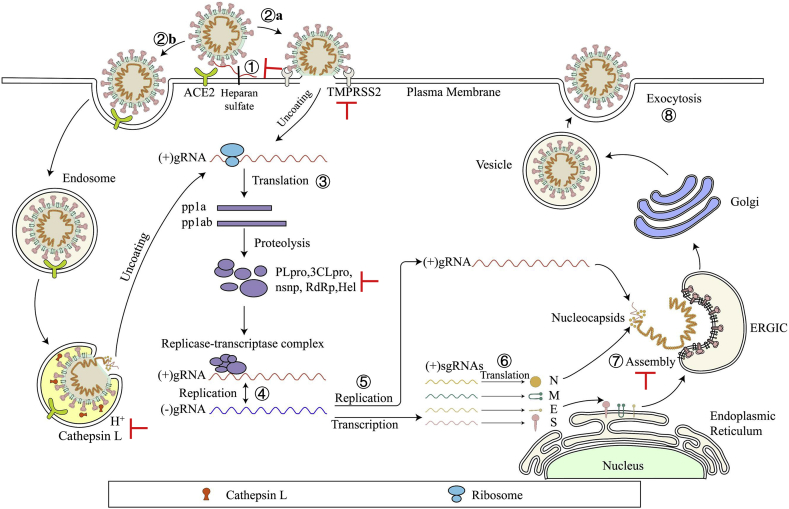
Figure 2.
Life cycle of SARS-CoV-2. SARS-CoV-2 first binds, via its S protein, to the receptor ACE2 on the target cell (①). Then, the virus must gain access to the host cell cytosol through plasma (②a) or endosomal membrane fusion (②b). This is assisted by activation of S protein by TMPRRS2 (②a) or cathepsin B/L (②b), followed by fusion of the viral and cellular membranes. The viral genome is released, uncoated and translated into viral replicase polyproteins pp1a and 1 ab (③), which are then cleaved into nonstructural proteins (nsps) by viral proteinases as papain-like protease (PLpro) and 3C-like protease (3CLpro). Many of these nsps as RNA-dependent RNA polymerase (RdRp) or Helicase (Hel) then assemble into the replicase–transcriptase complex which replicates the (+)-sense genomic RNA ((+) gRNA). (−)-sense genomic RNA ((−) gRNA) is synthesized and used as a template to form (+) gRNA) and subgenomic RNAs (sgRNAs) (⑤). The viral structural proteins, S, E, and M are translated from sgRNAs (⑥) and inserted into the endoplasmic reticulum (ER), from where they are transported to the ER–Golgi intermediate compartment (ERGIC) to interact with the (+) gRNA-encapsidated N proteins and assemble into viral particles (⑦). The budded vesicles containing mature viral particles are then transported to the cell surface for release after maturation in the Golgi bodies (⑧). Possible targets for inhibitors are marked in red.