Abstract
Antimicrobial resistance (AMR) has been recognized as one of the greatest global threats for human and animal health. The present review retrieved up to date information on the epidemiology of AMR in the animal-source food chain in Ethiopia focusing on AMR in bacterial species isolated from food handlers, live animals, foods (animal origin and non-animal origin), and in environmental samples. Accordingly, pooled prevalence of AMR in the different sources was estimated. For data analysis, we used random effect meta-analysis and in order to avoid exclusion of studies with zero prevalence of antimicrobial resistance, Freeman-Tukey double arcsine transformation was applied. We identified 152 eligible studies and retrieved 4097 data records (183 in food handlers, 2055 in foods, 1040 in live animals and 819 for environmental samples) which together reported a total of 86,813 AMR tests with 64 different antimicrobial disks for 81 bacteria species. We present the pooled prevalence of AMR for major bacterium-antibiotic combination in different sample types. The pooled prevalence of AMR in bacteria from food producing live animals was 20%. High estimates of AMR pooled prevalence were found in bacteria identified from milk, food handlers and the environmental samples with 29%, and 28% in meat. In foods of non-animal origin, the prevalence was lower with 13%. In milk, the highest AMR estimate was found for penicillin (69%) followed by amoxicillin (51%). Regarding multi-drug resistance (MDR), the overall pooled prevalence was 74% among AMR positive samples. Microbes reported having a higher MDR pattern were: Staphylococcus spp. (96%), Salmonella spp. (81%) and Escherichia coli (77%). The present review revealed a high resistance against commonly used drugs for animal and human treatments and/or prophylaxis. In conclusion, the high estimate of prevalence of AMR observed in bacteria recovered from different sample sources related to the animal-source food chain (food, live animal and environment) can highlight the possible linkage among them. The MDR levels in several bacteria species are a clear indication that the threat is directed to many antimicrobials. Our review demonstrated that the high overall AMR resistance levels call for effective policy and intervention measures, which best address the problem along the food chain through a One Health approach.
Keywords: Systematic review, Meta-analysis, Ethiopia, AMR, Prevalence, MDR, One health
Highlights
-
•
Meta-analysis of antimicrobial resistance conducted from One Health perspectives.
-
•
Pooled prevalence of AMR for bacterium-antibiotic combination in different sample was reported.
-
•
Comparable high AMR prevalence estimates were found from milk, food handler and the environment.
-
•
MDR prevalence was 74%, and higher estimate in E. coli, Staphylococcus, Salmonella and Shigella.
1. Introduction
Antimicrobial resistance (AMR) has been recognized as one of the major health threats to people and animals of global concern impacting both developed and developing countries. AMR is a problem across national borders, different degrees of healthcare sophistication or national economic status [1]. It is predicted that the economic loss associated with AMR will increase dramatically by the mid-21st century [2]. Therefore, AMR is among the top challenges in achieving the 2030 UN Sustainable Development Goals (SDGs) [[2], [3], [4]]. Many microbes in any natural product in the biosphere can carry AMR genes and the extent of the AMR problem is only recently being understood [5]. The problem associated with AMR is multi-dimensional in the sense that it affects biological, economic, ecologic, social and development dimensions and accordingly Butaye et al. [6] described AMR problem as “a highly multifaceted topic at the interface of human, animal and plant health, food hygiene and environmental science”.
Though the factors affecting the occurrence of AMR are complex without doubt AMR is also linked to irrational or mis-use of antimicrobial agents (more specifically antibiotics) in humans, agriculture, and veterinary medicines coupled with the self-antigenic re-engineering capacity of microorganisms. Prescott (2014) [7] described the complexity and widespread nature of AMR as: “resistance anywhere is resistance everywhere”. There is evidence of multiple links between the human, animal and wider environmental that allow not only movement of the bacteria but also movement of the mobile genetic elements (MGEs) of microorganisms and thus contributing to the further spread of AMR [8]. As a result, various countries have implemented policies like introduction monitoring systems of resistance in food animals considering the public health risks of possible transfer of resistant bacteria or genes from animals or environment to humans [9,10].
Like disease surveillance, AMR surveillance provides evidence to inform decisions on interventions. For example, AMR surveillance can help in the impact evaluation of interventions on guidelines on antimicrobial usage or infection control. Moreover, presence of functional surveillance mechanisms can provide useful information on AMR emergence and occurrence for decision making [1]. Thus, understanding the epidemiology of AMR is key to developing effective strategies to target a reduction in the emergence and spread of AMR. Unfortunately, routine surveillance of AMR is absent in most low- and middle-income countries and surveillance capacity for AMR is inadequate in most African countries [11].
High rates of resistance among antimicrobials frequently used to treat common bacterial infections have been observed in East Africa as reported by the Global Antimicrobial Resistance and Use Surveillance System (GLASS) [12]. For instance, in E. coli and Klebsiella pneumoniae recovered from urine and blood specimen of the community and unknown origin, the rate of resistance to ciprofloxacin, an antimicrobial medicine commonly used to treat urinary tract infections, were 67.4% and 59.3% (Ethiopia), 63.5% and 36.2% (Sudan) and 47.5% and 16.7% (Uganda) respectively [13]. Thus, organized multi-sectorial, collaborative surveillance of AMR is a key for decision-making at national, regional and international level. It assists tracking the burden of AMR, trends in resistance to ensure that countries can design cost effective, evidence-based AMR response strategies [14,15].
In Ethiopia, some studies also showed increasing levels of AMR [[16], [17], [18], [19]]. Different studies in human patients, food animals, foods and the environment indicated the danger of losing worthy therapeutics. However, as the studies are not comprehensive and difficult to compare, it can be difficult to get the full picture of the problem and compilation of the current information is needed to properly assess the situation and gear our actions and limited resources towards the gaps. The complexity of AMR clearly demands for more holistic approaches which consider linkages within a system, such as for example the food chain, which a One Health approach is well placed to achieve as it promotes collaboration across sectors and disciplines.
There are few meta-analyses on AMR in Ethiopia had limited scope and coverage and focused on single microorganisms isolated from either human cases or foods of animal origin only. However, to the best of our knowledge, no systematic review and meta-analysis has been conducted from a One Health perspective looking at the human-animal-environment interface. The present review aimed at retrieving and analyzing the existing information on AMR in Ethiopia focusing on bacteria species in the animal-source food chain by including food handlers, live animals, foods (animal origin and non-animal origin) and the environment.
2. Methods
2.1. Literature search and retrieval
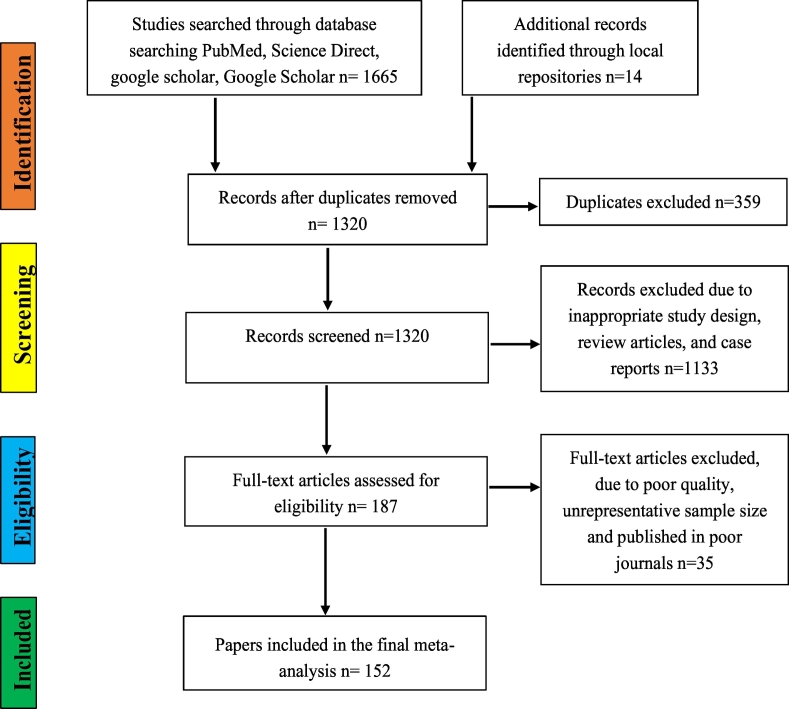
Systematic searching of various electronic databases such Google Scholar, Science Direct, PubMed, African Journals Online (AJOL) and Cab abstracts was conducted to retrieve relevant published articles. Online library repositories of different institutions were also searched. The process of retrieving and including data closely followed PRISMA guidelines [20] (Fig. 1). Relevant MeSH terms and keywords were used to retrieve all relevant articles from the above-listed databases. The keywords and MeSH terms used were: “AMR/antibiotic and Ethiopia”, “AMR/antibiotic prevalence and Ethiopia”, “E. coli and Ethiopia”, “Salmonella and Ethiopia”, “Staphylococcus and Ethiopia”, “Listeria and Ethiopia”, Pasteurella and Ethiopia”, “AMR/antibiotic in animals and Ethiopia”, “AMR/antibiotic prevalence in animals/livestock and Ethiopia”, “E. coli in animals and Ethiopia”, “Salmonella in animals/livestock and Ethiopia”, “Staphylococcus in animals/livestock and Ethiopia”, “Pasteurella in animals/livestock and Ethiopia”, AMR/antibiotic in foods and Ethiopia”, “AMR/antibiotic prevalence in foods and Ethiopia”, “E. coli in foods and Ethiopia”, “Salmonella in foods and Ethiopia”, “Staphylococcus in foods and Ethiopia”, “Listeria in foods and Ethiopia”, “AMR/antibiotic from water and Ethiopia”, “E. coli from water and Ethiopia”, “Salmonella water and Ethiopia”, “AMR/antibiotic from equipment and Ethiopia”, “E. coli from equipment and Ethiopia”, “Salmonella from equipment and Ethiopia”, and “Staphylococcus from equipment and Ethiopia”.
Fig. 1.
Flowchart of literature search and inclusion/exclusion process.
2.2. Inclusion criteria and exclusion criteria
The literature search defined by the above strategy returned a total of 1679 articles and for the identification of eligible articles we used pre-defined inclusion and exclusion criteria. The inclusion criteria were: 1) abstracts and full text available only in English 2) observational studies (cross-sectional/longitudinal) that report the proportion of bacterial pathogens and AMR, 3) sound methods for the detection of bacteria species (phenotypic identification using culture and biochemical test, serological or molecular detection methods), 4) similarly sound method in the assessment of AMR using either disk diffusion, E-test, minimum inhibitory concentration (MIC) inhibition or molecular techniques in compliance with international manuals and cut-off standards for AMR, 5) the source of samples for the bacteria isolation studies which should be from food handlers, live food animals, environmental samples, foods (animal origin or non-animal source foods), and 6) adequate representative sample size, and with a clear report on proportion of resistant and susceptible organisms. Accordingly, studies that did not comply with the above criteria were excluded. In addition, articles were not included when AMR information was missing, the studies aggregated AMR results into large categories such as “Gram-negative organisms,” and lacked the required full and clear information such as types of samples from which the bacteria were isolated, and studies conducted outside of Ethiopia.
2.3. Data extraction
Required information from the papers was extracted by four of the authors and then further verified for consistency and quality of the data. Data extraction templates were prepared in excel and pre-tested before the full extraction by extracting random papers by the team after which necessary adjustments were made. The variables extracted included the author name, year of publication, production system (especially for studies focusing on live animals), sample sources (food handlers, foods, food producing animals, and environment), sample size (number of study units/samples), number of positives (to calculate prevalence) and species of microorganism investigated, microbiological and AMR detection methods, for which antimicrobials were tested, number of susceptible and resistant pathogens, name of susceptible and resistant pathogens, and the presence of multidrug resistance (MDR) by pathogens detected. MDR was defined as resistance to two or more drugs in different antimicrobial classes in the context of this present review.
2.4. Data analysis
2.4.1. Descriptive summary
The results were summarized by presenting sample source/type, number of studies, sample size, number, and species of bacteria isolated and tested for AMR, the types of antimicrobials for the bacteria species tested and the overall prevalence of AMR or MDR.
2.4.2. Meta-analysis
The meta-analysis was carried out using Stata 14 software. The prevalence of AMR was pooled to estimate according to the different sources from which bacteria were isolated. The pooled AMR prevalence was reported if at least three different studies reported on a specific bacterium-antibiotic combination. We used Freeman -Tukey Double Arcsine Transformation for the data given that many zero prevalence of antimicrobial resistance were observed. The transformation helped to avoid exclusion of studies from the meta-analysis because of zero values. The total number of isolates tested for AMR and the number of resistant organisms were used to generate effect size (ES) and standard error (SE) along with their 95% confidence intervals. After that, the ES and SE were used to pool estimates of AMR levels [21]. In order to account for the heterogeneity in the data (expected to be high), random-effects meta-analysis was used to pool the estimates following procedure described by Higgins and Thompson (2002) [22]. As an additional procedure to minimize heterogeneity and to observe variations in AMR in different sample types, the meta-analysis was carried out by disaggregating the data according to the types of samples from which bacteria species were isolated (i.e. separately for food handlers, live animals and foods). In each sampling unit, repeatedly tested antimicrobial disks were prioritized for the meta-analysis to estimate the level of resistance for each antimicrobial compound.
3. Results
3.1. Descriptive summary of studies included
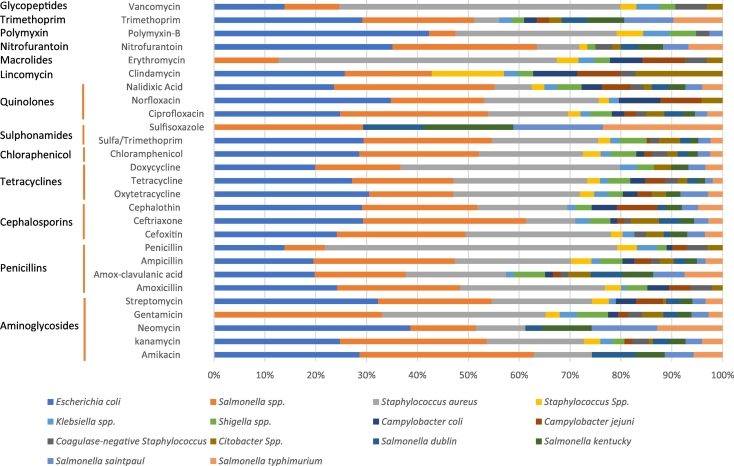
This systematic review and meta-analysis included 152 eligible studies. From these studies, we extracted 4097 records/data points, representing reports of 86, 813 AMR tests with different disks and microorganisms in food handlers, animals, foods and the environment. The included articles had tested for resistance to 64 different antimicrobial drugs involving 81 different micro-organisms. The drug classes and major antimicrobial drugs assessed are shown in Fig. 2. The characteristics of sample type, number of samples collected, type and proportion of bacterial species isolated and tested along sample source are described in supplementary appendix Table A.1.
Fig. 2.
Proportion of major species bacteria tested for major type of antibacterial disks and drug class.
3.2. Meta-analysis
The grouping of sample sources followed for the presentation of the results was food producing live animals (mostly with cases of mastitis, septicemia, and diarrhea), milk and milk products, meat and meat products, foods of non-animal origin, eggs, food handlers, and environmental samples (include swabs from swage systems, abattoir environment, water, and working utensils). Results of the meta-analysis were also presented by further disaggregating into various subgroups based on the antimicrobial disks and livestock production systems.
3.2.1. Prevalence of antimicrobial resistance in food-producing live animals
A total of 28 studies with 1040 data points reported AMR on live animals from clinical samples (mastitis, urine, feces, blood and other samples), farms clinics and household surveys. The pooled prevalence of AMR for bacteria isolated from samples of live animals was 20% (95% CI: 15%–24%), equivalent to one in five tests carried out revealed resistance to antimicrobial agents. The most commonly tested bacteria and estimate of AMR are depicted in a supplementary material (appendix Table A.2). High estimates of resistance were observed in Salmonella kentucky, Pasteurella multocida and Pasteurella haemolytica.
The result of subgroup analysis by antimicrobial disk tested showed that the most common resistances recorded were against streptomycin, ampicillin and tetracycline (Table 1). Accordingly, high level of pooled resistance prevalence of oxytetracycline 79% (95% CI 32% - 100%) was observed for E. coli isolated from animal samples and susceptible to gentamicin 0% (0% - 10%). For Salmonella spp., a higher level of resistance was observed for streptomycin 49% (33% - 66%), ampicillin 41% (25% - 57%), and oxytetracycline 44% (29%–58%) compared to cefoxitin (5%), ciprofloxacin (4%), and gentamicin (1%) (Table 2).
Table 1.
Subgroup analysis by antimicrobial disks and pooled prevalence of AMR in bacteria isolated from samples of live food animals in Ethiopia.
| Antimicrobials tested | Pooled prevalence of AMR (95% CI) | Heterogeneity |
|||
|---|---|---|---|---|---|
| I2% | Q | DF | P-Value | ||
| Streptomycin | 42% (30% - 54%) | 89.2 | 732.5 | 79 | 0.01 |
| Ampicillin | 36% (24% - 47%) | 83.5 | 432.5 | 71 | 0.01 |
| Tetracycline | 33% (20% - 47%) | 91.5 | 617.8 | 52 | 0.01 |
| Kanamycin | 23% (9% - 39%) | 85.7 | 372.0 | 53 | 0.01 |
| Sulfamethoxazole/Trimethoprim | 16% (5% - 29%) | 85.2 | 353.3 | 52 | 0.01 |
| Nalidixic Acid | 12% (2% - 25%) | 89.0 | 485.2 | 53 | 0.01 |
| Ciprofloxacin | 9% (1% - 21%) | 92.3 | 770.4 | 59 | 0.01 |
| Gentamicin | 9% (1% - 20%) | 90.4 | 723.9 | 69 | 0.01 |
| Chloramphenicol | 9% (1% - 18%) | 89.7 | 760.1 | 78 | 0.01 |
| Overall | 20% (16% - 24%) | 89.9 | 5695 | 574 | 0.01 |
Table 2.
The pooled prevalence of AMR for major bacterium-antibiotic combination in different sample types in Ethiopia.
| Sample type | Major bacteria identified and tested | TE |
S |
GEN |
K |
AMP |
AMC |
AMO |
P |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | P (95% CI) | N | P (95% CI) | N | P (95% CI) | N | P (95% CI) | N | P (95% CI) | N | P (95% CI) | N | P (95% CI) | N | P (95% CI) | ||
| Food animal | E. coli | 4 | 79 (32−100) | 5 | 31 (5–65) | 3 | 0 (0−10) | ||||||||||
| Salmonella spp. | 9 | 44 (29–58) | 10 | 49 (33–66) | 8 | 1 (0–6) | 9 | 17 (4–35) | 9 | 41 (25–57) | 5 | 17 (6–31) | 4 | 78 (61–91) | |||
| Pasteurella haemolytica | 6 | 28 (13–48) | 6 | 54 (29–79) | 4 | 81 (51–100) | 4 | 47 (22–72) | |||||||||
| Campylobacter jejuni | 4 | 21 (2–50) | 3 | 9 (5–14) | |||||||||||||
| Milk | E. coli | 11 | 44 (20–70) | 12 | 49 (17–43) | 12 | 15 (3–34) | 9 | 72 (49–91) | 4 | 56 (14–94) | 5 | 51 (10–91) | ||||
| Salmonella spp. | 8 | 66 (42–87) | 6 | 27 (6–56) | 10 | 3 (0−13) | 5 | 75 (43–98) | 5 | 55 (8–97) | |||||||
| Staphylococcus aureus | 28 | 45 (36–55) | 17 | 31 (20–43) | 23 | 6 (2−11) | 16 | 8 (1–17) | 17 | 60 (44–75) | 6 | 8 (0−21) | 10 | 53 (36–69) | 27 | 82 (74–89) | |
| Streptococcus agalactiae | 4 | 46 (31–61) | 4 | 26 (8–49) | 3 | 4 (0−12) | 4 | 51 (24–77) | 3 | 61 (47–74) | 4 | 54 (14–92) | 3 | 53 (21–83) | |||
| Meat | E. coli | 12 | 38 (21–56) | 10 | 32 (16–49) | 6 | 32 (7–63) | 8 | 35 (3–75) | 5 | 77 (64–88) | 6 | 76 (40–100) | ||||
| Salmonella spp. | 10 | 55 (29–80) | 9 | 60 (31–87) | 12 | 2 (0–9) | 9 | 23 (2–51) | 13 | 38 (15–63) | 5 | 58 (29–85) | 5 | 27 (0–72) | |||
| Campylobacter spp. | 3 | 14 (4–28) | 3 | 12 (3–24) | |||||||||||||
| Staphylococcus aureus | 5 | 29 (0–75) | 3 | 64 (22–98) | |||||||||||||
| FNA | E. coli | 6 | 14 (0–55) | 5 | 8 (0−32) | 5 | 78 (60–92) | 3 | 16 (0–81) | ||||||||
| Salmonella Spp. | 6 | 20 (6–37) | 3 | 30 (3–67) | 5 | 0 (0–1) | 5 | 96 (77–100) | |||||||||
| Staphylococcus aureus | 4 | 30 (4–64) | 3 | 26 (0–85) | 5 | 4 (0–13) | 4 | 61 (0−100) | 3 | 78 (31−100) | |||||||
| Food handler | Salmonella Spp. | 3 | 39 (4–81) | 3 | 0 (0–17) | 5 | 90 (76–99) | 3 | 32 (7–63) | ||||||||
| Shigella Spp. | 4 | 74 (55–90) | 3 | 18 (5–35) | 4 | 82 (61–97) | |||||||||||
| Staphylococcus spp. | 3 | 46 (25–68) | |||||||||||||||
| Envr. | E. coli | 9 | 21 (4–45) | 7 | 13 (2–29) | 7 | 97 (86–100) | 5 | 51 (18–84) | 3 | 71 (12−100) | ||||||
| Staphylococcus aureus | 5 | 34 (5–72) | 4 | 3 (0–17) | 4 | 31 (0–91) | 5 | 89 (73–99) | |||||||||
| Klebsiella spp. | 5 | 33 (11–58) | 5 | 10 (4–19) | 5 | 70 (35–96) | 5 | 34 (10–63) | |||||||||
| Enterobacter spp. | 3 | 23 (11–37) | 3 | 11 (2–24) | 3 | 57 (30–82) | |||||||||||
| Salmonella spp. | 3 | 65 (44–83) | 4 | 4 (0–27) | |||||||||||||
| Pseudomonas spp. | 3 | 86 (46–100) | 3 | 23 (2–52) | 3 | 100 (86–100) | 3 | 96 (77–100) | |||||||||
| Sample type | Major bacteria identified and tested | CAP |
CTX |
CIP |
NA |
NOR |
SXT |
ERY |
CRO |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | P (95% CI) | N | P (95% CI) | N | P (95% CI) | N | P (95% CI) | N | P (95% CI) | N | P (95% CI) | N | P (95% CI) | N | P (95% CI) | ||
| Food animal | E. coli | 4 | 19 (0–75) | 3 | 41 (0–97) | ||||||||||||
| Salmonella spp. | 10 | 9 (1−21) | 5 | 5 (0−30) | 9 | 4 (0–12) | 7 | 16 (3−32) | 7 | 10 (0–28) | |||||||
| Pasteurella haemolytica | 6 | 0 (0–34) | |||||||||||||||
| Campylobacter jejuni | 3 | 10 (0–50) | 4 | 18 (2–42) | |||||||||||||
| Milk | E. coli | 9 | 34 (14–58) | 8 | 0 (0–3) | 5 | 18 (4–39) | 6 | 32 (9–61) | ||||||||
| Salmonella spp. | 6 | 30 (19–43) | 8 | 3 (0–13) | 7 | 41 (6–81) | 6 | 8 (0–27) | |||||||||
| Staphylococcus aureus | 22 | 11 (3−21) | 12 | 36 (22–52) | 8 | 8 (0–26) | 5 | 52 (14–89) | 3 | 0 (0–7) | 17 | 21 (9–38) | 24 | 12 (6–19) | |||
| Streptococcus agalactiae | 3 | 26 (0–72) | |||||||||||||||
| Meat | E. coli | 9 | 39 (18–62) | 4 | 58 (13–97) | 5 | 3 (0–13) | 8 | 34 (5–71) | 4 | 33 (1–76) | 9 | 24 (5–49) | 3 | 45 (23–68) | ||
| Salmonella spp. | 11 | 4 (0–18) | 5 | 22 (2–52) | 10 | 0 (0–5) | 10 | 9 (0–29) | 9 | 9 (0−23) | 6 | 4 (0–19) | |||||
| FNA | E. coli | 6 | 5 (0–14) | 5 | 10 (0–43) | 3 | 17 (0–59) | 3 | 17 (0–54) | 3 | 0 (0–2) | ||||||
| Salmonella Spp. | 6 | 9 (0–36) | 6 | 0 (0–1) | 4 | 40 (5–81) | 4 | 0 (0–2) | 3 | 23 (0–84) | |||||||
| Staphylococcus aureus | 5 | 12 (1−32) | 5 | 1 (0–9) | 3 | 0 (0–1) | 3 | 8 (0–46) | 4 | 19 (0–58) | |||||||
| Food handler | Salmonella Spp. | 4 | 16 (7–27) | 5 | 6 (1–14) | 3 | 50 (31–69) | 3 | 19 (9–31) | ||||||||
| Shigella Spp. | 4 | 27 (2–62) | 4 | 1 (0–13) | 3 | 7 (0–19) | 4 | 32 (10–59) | |||||||||
| Envr. | E. coli | 7 | 17 (3–37) | 4 | 14 (0–41) | 7 | 12 (0−31) | 9 | 17 (4–35) | 3 | 0 (0–1) | 4 | 21 (1–53) | ||||
| Staphylococcus aureus | 5 | 30 (4–64) | 4 | 6 (2−12) | 5 | 11 (0–39) | 5 | 18 (1–45) | |||||||||
| Klebsiella spp. | 3 | 25 (7–49) | 5 | 11 (0–32) | 5 | 30 (15–48) | 4 | 26 (16–37) | |||||||||
| Enterobacter spp. | 3 | 23 (14–34) | 3 | 11 (0–36) | 4 | 30 (15–47) | |||||||||||
| Salmonella spp. | 3 | 41 (0–95) | 4 | 33 (1–77) | |||||||||||||
| Pseudomonas spp. | 3 | 70 (19–100) | 3 | 0 (0–14) | 3 | 70 (19–100) | |||||||||||
FNA Food of non-animal origin, Envr. Environment, N Number of studies, P Pooled prevalence of AMR, CAP Chloramphenicol, TE Tetracycline, S Streptomycin, GEN Gentamicin, K Kanamycin, CTX Cefoxitin, AMP Ampicillin, AMC Amoxicillin-clavulanic acid, AMO Amoxicillin, P Penicillin, CIP Ciprofloxacin, NA Nalidixic Acid, NOR Norfloxacin, SXT Sulfamethoxazole/Trimethoprim, ERY Erythromycin, CRO Ceftriaxone.
The results of subgroup analysis by livestock production system indicated that the AMR prevalence differed across three production systems (Table 3).
Table 3.
Subgroup meta-analysis by production systems and the pooled prevalence of AMR in food animals in Ethiopia.
| Production system | Pooled prevalence of AMR (95% CI) | Heterogeneity |
||
|---|---|---|---|---|
| I2% | Q | P-Value | ||
| Peri-urban | 27% (20% - 34%) | 87.8 | 1253.70 | 0.01 |
| Pastoralist | 23% (7% - 44%) | 98.3 | 645.53 | 0.01 |
| Urban | 17% (13% - 22%) | 87.6 | 3303.45 | 0.01 |
| Overall | 20% (16% - 24%) | 89.92 | 5695.32 | 0.01 |
3.2.2. Prevalence of antimicrobial resistance in milk and milk products
A total of 933 data points was extracted from a total of 53 studies which involved bacteriological analysis of milk samples with the tested microorganisms showed an overall pooled prevalence of resistance of 29% (95% CI: 25–33%, and heterogeneity chi-square value 10,812.40 (p-value of 0.001), I2 = 95.37% and an estimate of between-study variance (τ2 = 0.61). The pooled resistance prevalence of specific bacterium-antibiotic combinations in milk are described in Table 2. A high prevalence estimate of AMR was observed in Staphylococcus spp. (70%) followed by Streptococcus agalactiae (53%) (Table A.1.2). The result of the pooled prevalence of AMR for different antibiotic disks (Table 4) indicated the highest level for resistance against penicillin, followed by amoxicillin and tetracycline.
Table 4.
Subgroup analysis of resistance against selected antimicrobials for bacteria isolated from milk and milk products.
| Antimicrobials tested | Pooled prevalence of AMR (95% CI) | Heterogeneity |
||
|---|---|---|---|---|
| I2% | Q | P-Value | ||
| Penicillin | 69% (58% - 79%) | 95.4 | 1458.1 | 0.01 |
| Amoxicillin | 51% (39% - 62%) | 88.0 | 368.4 | 0.01 |
| Tetracycline | 43% (34% - 52%) | 92.4 | 922.7 | 0.01 |
| Streptomycin | 27% (20% - 34%) | 90.7 | 732.2 | 0.01 |
| Erythromycin | 22% (15% - 29%) | 92.4 | 652.5 | 0.01 |
| Chloramphenicol | 19% (11% - 28%) | 94.6 | 1218.3 | 0.01 |
| Kanamycin | 16% (6% - 26%) | 93.3 | 868.4 | 0.01 |
| Gentamycin | 6% (3% - 10%) | 87.6 | 599.7 | 0.01 |
| Overall | 29% (25% - 33%) | 95.3 | 10,812.4 | 0.01 |
The analysis of AMR pooled prevalence in milk and milk products by production system found for pastoralist areas a prevalence of 42% (22% - 64%), followed by urban systems with 29% (24% - 35%) and peri-urban systems with 28% (23% - 33%).
3.2.3. Prevalence of antimicrobial resistance in meat
A total of 37 studies with 763 data points were extracted from studies on AMR in meat. The pooled prevalence of AMR of bacteria in meat product samples was 28% (95% CI: 24–32%). High prevalence estimates of AMR were observed in E. coli (41%), Staphylococcus aureus (30%) and Salmonella spp. (28%) (Table A.1.2). AMR prevalence was pooled for the antimicrobials used in the studies through subgroup analysis for each drug. The highest levels were found for ampicillin, tetracycline and streptomycin (Table 5). E. coli showed high level of resistance for most of the antimicrobials, with the highest resistance against ampicillin (76%) and the lowest against ciprofloxacin (3%). Among Salmonella spp. isolates, the lowest pooled AMR prevalence levels were found for ciprofloxacin (0%), gentamicin (2%), chloramphenicol (4%), and ceftriaxone (4%). The upper levels of AMR were recorded for other drugs such as streptomycin (60%), amoxicillin-clavulan acid (58%), and tetracycline (55%) (Table 2). For meat samples, further subgroup analysis by production systems indicated that the pooled prevalence of AMR in urban production system was 24% (20% - 29%), while it was 34% (28% - 41%) in peri-urban production system.
Table 5.
Common antimicrobial drugs and corresponding AMR prevalence for bacteria isolated from meat in Ethiopia.
| Antimicrobials tested | Pooled prevalence of AMR (95% CI) | Heterogeneity |
||
|---|---|---|---|---|
| I2% | Q | P-Value | ||
| Ampicillin | 40% (25–56%) | 92.9 | 656.17 | 0.001 |
| Tetracycline | 37% (24%–50%) | 92.9 | 858.56 | 0.001 |
| Streptomycin | 32% (21%–44%) | 87.7 | 416.4 | 0.001 |
| Nalidixic Acid | 25% (11%–43%) | 93.3 | 615.66 | 0.001 |
| Kanamycin | 23% (10%–39%) | 91.4 | 468.38 | 0.001 |
| Sulfamethoxazole/Trimethoprim | 16% (7%–27%) | 87.4 | 366.55 | 0.001 |
| Gentamycin | 10% (3–18%) | 89.0 | 355.44 | 0.001 |
| Overall | 28% (24%–32%) | 93.6 | 11,990.21 | 0.01 |
3.2.4. Prevalence of antimicrobial resistance in eggs
Four studies with a total of 65 records reported AMR in egg samples with an overall AMR prevalence of 36% (95% CI: 21%–53%). The most commonly isolated bacteria were Salmonella spp. with estimate of AMR of 34% (95% CI: 16%–54%). The prevalence of tetracycline resistance of organisms was estimated 93% (95% CI: 67–100%). Moreover, the overall pooled prevalence of amoxicillin resistance was 51% (95% CI: 1–99%). Further subgroup analysis by production system indicated that the pooled prevalence AMR in urban production systems was 42% (95% CI: 21–64%) while it was 29% (95% CI: 16–35%) in peri-urban production system.
3.2.5. Prevalence of antimicrobial resistance in foods of non-animal origin
Ten studies resulting in 294 data points investigated AMR prevalence in restaurant served foods, street foods and university restaurant foods with an overall pooled prevalence of AMR in bacteria identified from foods of non-animal origin was 13% (95% CI: 10%–18%). High AMR estimate was observed in Staphylococcus aureus and E. coli (20% for both) compared to Salmonella spp. (16%) (Table A.1.2). Subgroup analysis indicated that highest prevalence was observed for ampicillin resistance, while the lowest prevalence was observed for gentamycin, streptomycin and kanamycin resistance (Table 6).
Table 6.
Subgroup analysis by antimicrobials and pooled resistance prevalence in foods of non-animal origin in Ethiopia.
| Antimicrobials tested | Pooled prevalence of AMR (95% CI) | Heterogeneity |
||
|---|---|---|---|---|
| I2% | Q | P-Value | ||
| Ampicillin | 73% (52% - 90%) | 95.4 | 417.2 | 0.01 |
| Tetracycline | 28% (11% - 49%) | 95.8 | 336.6 | 0.01 |
| Penicillin | 22% (1% - 55%) | 95.0 | 242.2 | 0.01 |
| Sulfamethoxazole/Trimethoprim | 16% (1% - 38%) | 90.5 | 127.3 | 0.01 |
| Erythromycin | 9% (0% - 25%) | 91.2 | 159.2 | 0.01 |
| Chloramphenicol | 6% (1% - 12%) | 81.6 | 131.0 | 0.01 |
| Ciprofloxacin | 2% (0% - 9%) | 85.1 | 134.7 | 0.01 |
| Kanamycin | 2% (0% - 10%) | 82.7 | 69.6 | 0.01 |
| Gentamicin | 1% (0% - 5%) | 74.6 | 86.8 | 0.01 |
| Overall | 13% (10% - 18%) | 94.8 | 3016.3 | 0.01 |
3.2.6. Prevalence of antimicrobial resistance in environmental samples
Ten studies investigated AMR in the environment (sewage systems, abattoir environment, water, and working utensils) resulting in 819 data points. The pooled prevalence of AMR in bacteria isolated from environmental samples was 29% (95 CI: 25% - 32%). Various bacteria were detected from the studies and the most commonly isolated bacteria and estimate of resistance was showed in Table A.1. 2. High AMR prevalence was observed in Pseudomonas aeruginosa (67%), Citobacter spp. (47%) and Acinetobacter spp. (44%). Subgroup analysis of AMR by antimicrobial disks indicated that the highest prevalence was observed against Ampicillin, followed by amoxicillin and nitrofurantoin (Table 7).
Table 7.
Subgroup analysis by antimicrobials and pooled resistance prevalence in environmental samples in Ethiopia.
| Antimicrobials tested | Pooled prevalence of AMR (95% CI) | Heterogeneity |
||
|---|---|---|---|---|
| I2% | Q | P-Value | ||
| Ampicillin | 82% (69% - 93%) | 90.5 | 517.6 | 0.01 |
| Amoxicillin | 62% (49% - 74%) | 83.4 | 211.5 | 0.01 |
| Nitrofurantoin | 55% (44% - 57%) | 80.6 | 175.9 | 0.01 |
| Amoxicillin-clavulanic | 39% (27% - 51%) | 78.6 | 182.2 | 0.01 |
| Penicillin | 33% (14% - 55%) | 95.2 | 593.1 | 0.01 |
| Tetracycline | 31% (21% - 41%) | 86.0 | 430.4 | 0.01 |
| Sulfamethoxazole/Trimethoprim | 24% (17% - 33%) | 83.3 | 455.2 | 0.01 |
| Chloramphenicol | 23% (14% - 34%) | 87.2 | 463.9 | 0.01 |
| Nalidixic Acid | 22% (9% - 38%) | 91.3 | 276.4 | 0.01 |
| Ciprofloxacin | 9% (4% - 16%) | 81.9 | 315.9 | 0.01 |
| Gentamicin | 9% (5% - 14%) | 60.8 | 150.8 | 0.01 |
| Erythromycin | 6% (1% - 13%) | 81.0 | 147.7 | 0.01 |
| Vancomycin | 4% (0% - 16%) | 91.7 | 325.3 | 0.01 |
| Overall | 29% (25% - 32%) | 90.1 | 5961.6 | 0.01 |
3.2.7. Prevalence of antimicrobial resistance in food handlers
Ten studies investigated AMR among food handlers resulting in 183 data points with 29% pooled prevalence of AMR of bacteria. The most commonly isolated bacteria and estimate of resistance is showed in Table A.1. 2. High estimates of AMR were observed in Salmonella spp. (37%), E. coli (36%) and Shigella spp. (35%).
The pooled prevalence of ampicillin resistance was 89% (95% CI: 81–95%). Furthermore, the prevalence of chloramphenicol resistance of organisms was estimated 15% (95% CI: 7–25% and Heterogeneity chi-square value of 44.66 and p = 0.00, I2 (variation in ES attributable to heterogeneity) = 59.70%, Estimate of between-study variance τ2 = 0.1). The analysis for chloramphenicol resistance had a reasonably lower variability between studies. Similarly, the pooled prevalence of tetracycline and sulfamethoxazole/trimethoprim resistance was 56% (95% CI: 41–71%) and 35% (95% CI: 23–48%), respectively.
3.2.8. Prevalence of multidrug resistance (MDR) pattern
The prevalence of MDR was pooled from studies that reported the presence of MDR. Sixty-five articles with 101 records were eligible for the MDR meta-analysis. The overall MDR pooled prevalence was 74%. Microbes reported having higher MDR pattern were Staphylococcus spp. other than Staphylococcus aureus followed by Salmonella spp. (Table 8).
Table 8.
Subgroup analysis of the prevalence of MDR by selected species of organisms in Ethiopia.
| Species of organism tested | Pooled prevalence of MDR (95% CI) | Heterogeneity |
||
|---|---|---|---|---|
| I2% | Q | P-Value | ||
| Staphylococcus spp. other than Staph. aureus | 96% (81% - 100%) | 63.7 | 8.2 | 0.04 |
| Salmonella spp. | 81% (66% - 93%) | 92.2 | 438.2 | 0.00 |
| E. coli | 77% (65% - 88%) | 90.0 | 291.8 | 0.00 |
| Shigella spp. | 68% (49% - 86%) | 63.6 | 19.2 | 0.01 |
| Staphylococcus aureus | 58% (44% - 72%) | 96.2 | 618.4 | 0.00 |
| Overall | 74% (67% - 81%) | 93.1 | 1460.5 | 0.00 |
4. Discussion
The global AMR threat to the health of humans and animals is real and has attracted global attention to tackle its spread before it is too late. AMR is usually not systematically monitored in under-resourced countries because of lack of surveillance networks, laboratory capacity, and appropriate diagnostics [23]. This is compounded by poor access to different drugs which results in mis-use or inadequate use of antibiotics.
This systematic review and meta-analysis on AMR in Ethiopia covered extensive literature with massive datasets. The datasets were from different sources with an enormous variability which was reflected by the resulting high heterogeneity between studies. Conducting the analysis by sample type groups helped to address the heterogeneity and allowed observing the pattern of antimicrobial resistance between sources from a One Health perspective.
In this study, the overall pooled prevalence of AMR in bacteria recovered from samples collected from live food animals was 20%. As administering antimicrobials in animals is a key contributor to AMR prevalence in these animals and is becoming a pressing issue, these findings are of concern and should be taken seriously as drug sensitivity tests are not carried out as part of the diagnostic process and therapy is empirical in many clinical settings of Ethiopia. This might lead to increased morbidity and mortality in animal population affected by the resistant pathogens due to the less effective antimicrobial treatment. A large proportion of the population in Ethiopia live in proximity with animals, thus there is a risk of transmission of resistant microorganisms (with resistance genes) from animals to humans through the consumption of food or through direct contact with food-producing animals or through environmental spread (e.g. human sewage and runoff water from agricultural sites) [24]. The comparably higher resistance prevalence observed in pastoralist settings can be due to the fewer studies included, or the practice of extensive antimicrobials usage due to higher animal population and widespread infectious diseases.
The overall pooled prevalence of AMR in bacteria recovered from milk and milk products was 29% with concerning levels of penicillin resistance in most organisms studied, calling for a halt of widespread use of this drug in the dairy sector. High levels of resistance were also observed in Staphylococcus spp. which may lead to difficulties in effectively treating mastitis in the future. The higher AMR prevalence in milk from pastoralists than the other production system can be due to extensive antimicrobial administration by CAHWs and potentially wrong use of antibiotics by pastoralists [25], or the fat that fewer studies were available for pastoralist systems which may have resulted in biased estimates with inflated prevalence.
In meat and meat products, ampicillin resistance in E. coli was notable and for Salmonella spp. the key resistances were against streptomycin, amoxicillin-clavulan acid, and tetracycline, reflecting the use patterns in these animals. However, a study conducted in 2019 noted 25% of Salmonella isolates was recorded resistance against ampicillin, 10% of E. coli O157:H7 isolates were resistant against ciprofloxacin, and the AMR profile of bacterial isolates from meat and meat products was found less than 10% in majority of estimates [26]. The lower estimate of AMR in the later study could be due to its limited scope in slaughterhouses and markets in urban area of the country where most abattoirs are located. Previous reports elsewhere also indicated many microbes have developed resistance to these drugs [27].
The lower AMR prevalence in foods of non-animal origin can be expected as the contact of antimicrobials and these food types are low except in cases of resistant genes transferring from other sources. The AMR prevalence in these sample groups ranged from 1% in gentamicin to 73% in amoxicillin, indicating amoxicillin is getting resisted by many microbes everywhere. Previous reports indicated these drugs are becoming less efficient in humans [28].
The higher estimates of resistance from the environmental samples shown like swage systems, abattoir environment, water, and working utensils in this study tells us that the environment can be a significant part in the resistance transmission scenario. This area (environment) is a key area where resistance genes from animals and humans flow to environmental pathogens, can be maintained and from where it can spread further [29].
Although data on occurrence of the AMR in wildlife is currently very limited in the country, wildlife may also play an important role in relation to antibiotic resistance in the One Health concept as part of the environment [30]. They have been recognized as possible reservoirs and highly influenced by human activities especially in countries with poor hygiene standards and sanitation, with insufficient wastewater treatment and waste management, where animals and humans live close together, and where clinically important multi-drug resistant bacteria are widely present in the environment [30,31].
The AMR in food handlers who have close contact with food or those who prepare food in university restaurants, public restaurants, and abattoir employees is also of concern considering the threat it poses to human health. Moderately higher AMR is noted among pathogens such as Salmonella spp., Shigella spp. and E. Coli to commonly prescribed antibiotics, including ampicillin and tetracycline compared to other categories of samples. Ampicillin is becoming useless amid many microbes from this source are developing resistance. Similarly, E. coli isolates recovered from wound infection exhibited the highest point estimate of resistance against ampicillin, 84% followed by amoxicillin, 73% [32]. Many reports have shown that many microbes are getting the ability to resist tetracycline effectively [28].
There is a high level of MDR bacteria in the country (74%) estimated in this study which would make empiric antibiotic use challenging. This is an indication that microbes can avoid the effect of more than two drugs. Staphylococcus spp. other than Staphylococcus aureus showed the highest rate of MDR with prevalence of 96%, followed by E. coli 77% and Salmonella spp. 81%. These organisms are the predominant organism well studied phenotypically in the country with widespread prevalence in foods and food-producing animals. Being exposed to wide range antimicrobials, they evolved with the potential to resist the effect of many antimicrobials available in the market. Comparably, pooled MDR (defined by resistance to at least two antibiotics) rate of 59.7% was noted in hospital setting in the country by Muhie [33]. MDR bacteria have been also detected in meat and fresh produce [34] and in humans in contact with livestock in many African countries [35,36]. Reports in Cameroon for selected microbes showed a higher MDR, which is in line with this finding [27].
Apart from the irrational use of antimicrobials, unique environmental conditions such as crowding and poor sanitation also contribute in the circulation and spread of resistant microorganisms. Transmission of resistant pathogens is facilitated by person-person contact, through contaminated water, food or by vectors. Improving basic hygiene and sanitation will reduce the spread of resistant organisms [24]. Given the widespread consumption practice of raw meat and milk in the country and poor hygienic management, especially in the rural community of the country, there is a high risk of human exposure of these MDR organisms. Hence, establishment and strengthening the National Antimicrobial Resistance Surveillance System is mandatory to properly understand and address the prevailing problem in the country. Microbial ecology and drug resistance pattern studies in Ethiopia is at its infantry stage. More advanced studies in terms of AMR gene detection and the flow pattern in the human-animal-environmental interface needs to get attention to tackle this threat in the country effectively.
5. Conclusion
This extensive systematic review and meta-analysis presented a summary of the current status of AMR in Ethiopia from One Health perspective.
The analysis clearly showed the overall drug resistance problem across different sample sources with higher estimates of clinically important resistance from the milk, meat, food handler and the environment. It also highlighted the need to understand the possible linkage, transmission pathways and flow pattern of resistant bacteria and the AMR genes in human-animal-environmental interface in order to minimize the potential hazard to public health. Therefore, interventions to reduce the burden of antimicrobial resistance should be grounded by such understanding of the essence of coordinated One Health approach.
AMR is becoming a serious health issue in the country as some widely used antimicrobials are becoming not effective in the treatment of bacterial infections in animals and humans. The highest drug resistance was noted in drugs like ampicillin, amoxicillin, streptomycin and tetracycline. Therefore, drug susceptibility for treating bacterial infections should be practiced as much as possible. On time revision and implementation of standard treatment guidelines shall be prioritized as strategy to tackle AMR in the country. The result of MDR is a clear indication that the treatment is directed to many antimicrobials by pathogenic organisms like Staphylococcus spp., E. coli, and Salmonella spp. which needs serious attention from responsible bodies before it is too late, and the country should consider AMR as a public health priority issue.
Ethical statement
Not applicable.
Funding
This work was funded by the Animal Health Flagship of the CGIAR Research Program on Livestock.
Authors' contributions
BA, KA, AA and BW conceived and designed the study, BA, AA, KA, MB acquired relevant literature and extracted the data. BA and AA analysed and interpreted the data and drafted the manuscript. BA, AA, KA, MB and BW conceptualized and critically reviewed the manuscript. All authors read, commented and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Not applicable.
Appendix A. Appendices
Table A.1.
Distribution of studies used, type and proportion of bacterial species isolated and tested along sample source.
| Sample source | No. of studies | Bacterial isolate | Sample size | No. of Positive (%) | No. test | No. resistant | No. of data points |
|---|---|---|---|---|---|---|---|
| Food handler | 10 | E. coli | 693 | 29(4.2) | 285 | 115 | 19 |
| Salmonella spp. | 2493 | 148(5.9) | 734 | 318 | 67 | ||
| Salmonella para typhi | 233 | 1(0.4) | 9 | 2 | 9 | ||
| Salmonella typhi | 233 | 2(0.9) | 18 | 3 | 9 | ||
| Shigella spp. | 1166 | 84(7.2) | 426 | 149 | 35 | ||
| Staphylococus spp. | 218 | 13(6) | 117 | 41 | 9 | ||
| Staphylococus aureus | 371 | 108(29.1) | 1364 | 300 | 23 | ||
| Campylobacter jejuni | 164 | 15(9.1) | 90 | 18 | 6 | ||
| Campylobacter coli | 164 | 1(0.6) | 6 | 1 | 6 | ||
| Animals | 28 | E. coli | 904 | 134(14.8) | 1350 | 348 | 46 |
| Salmonella spp. | 2958 | 285(9.6) | 3176 | 1020 | 190 | ||
| Salmonella typhimurium | 2112 | 14(0.7) | 257 | 35 | 43 | ||
| Salmonella dublin | 1929 | 13(0.7) | 207 | 48 | 30 | ||
| Salmonella virchow | 1563 | 9(0.6) | 202 | 44 | 63 | ||
| Salmonella saintpaul | 2838 | 32(1.1) | 494 | 140 | 61 | ||
| Salmonella braenderup | 1086 | 3(0.3) | 50 | 14 | 33 | ||
| Salmonella haifa | 1635 | 5(0.3) | 82 | 29 | 49 | ||
| Salmonella kottbus | 726 | 1(0.1) | 17 | 6 | 17 | ||
| Salmonella kentucky | 2838 | 12(0.4) | 178 | 89 | 62 | ||
| Salmonella mikawasima | 1929 | 2(0.1) | 30 | 7 | 30 | ||
| Salmonella indiana | 360 | 4(1.1) | 64 | 14 | 16 | ||
| Salmonella chailey | 360 | 1(0.3) | 16 | 6 | 16 | ||
| Salmonella anatum | 360 | 2(0.6) | 32 | 6 | 16 | ||
| Salmonella minnesota | 360 | 1(0.3) | 16 | 2 | 16 | ||
| Salmonella muenchen | 360 | 1(0.3) | 16 | 1 | 16 | ||
| Salmonella tarshyne | 360 | 1(0.3) | 16 | 1 | 16 | ||
| Salmonella livingstoneVar.14+ | 1203 | 1(0.1) | 13 | 1 | 13 | ||
| Salmonella aberdeen | 1203 | 1(0.1) | 13 | 1 | 13 | ||
| Salmonella bronx | 360 | 7(1.9) | 112 | 18 | 16 | ||
| Salmonella newport | 360 | 6(1.7) | 96 | 28 | 16 | ||
| Salmonella enterica subsp.enterica | 150 | 4(2.7) | 14 | 0 | 1 | ||
| Staphylococus aureus | 481 | 43(8.9) | 407 | 154 | 39 | ||
| Staphylococus spp. | 70 | 14(20) | 154 | 24 | 11 | ||
| Campylobacter jejuni | 1140 | 411(36.1) | 4920 | 1644 | 44 | ||
| Campylobacter coli | 780 | 90(11.5) | 903 | 118 | 27 | ||
| Campylobacter lari | 192 | 6(3.1) | 69 | 30 | 4 | ||
| B. anthracis | 70 | 2(2.9) | 22 | 1 | 11 | ||
| Mannheimia/Pasteurella haemolytica | 1099 | 133(12.1) | 1126 | 558 | 60 | ||
| Pasteurella multocida | 789 | 30(3.8) | 142 | 65 | 14 | ||
| Bibersteinia trehalosi | 149 | 7(4.7) | 49 | 20 | 7 | ||
| Streptococcus spp. | 70 | 2(2.9) | 22 | 1 | 11 | ||
| Pseudomonas aeruginosa | 70 | 1(1.4) | 11 | 10 | 11 | ||
| Listeria monocytogenes | 70 | 3(4.3) | 33 | 2 | 11 | ||
| Listeria spp. | 70 | 2(2.9) | 22 | 1 | 11 | ||
| Food (Milk) | 53 | Staphylococus aureus | 10,719 | 1998(18.6) | 21,100 | 7052 | 379 |
| Staphylococcus epidermidis | 1877 | 57(3) | 485 | 118 | 35 | ||
| Staphylococcus haemolyticus | 383 | 1(0.3) | 2 | 2 | 2 | ||
| Staphylococcus hyicus | 18 | 1(5.6) | 5 | 1 | 5 | ||
| Staphylococcus intermedius | 704 | 11(1.6) | 58 | 8 | 16 | ||
| Staphylococcus lentus | 383 | 2(0.5) | 4 | 2 | 2 | ||
| Staphylococcus xylosus | 383 | 4(1) | 8 | 6 | 2 | ||
| Staphylococcus sciuri | 383 | 3(0.8) | 6 | 2 | 2 | ||
| Staphylococus spp. | 1182 | 50(4.2) | 325 | 211 | 41 | ||
| CNS | 643 | 75(11.7) | 649 | 287 | 23 | ||
| MRSA | 384 | 110(28.6) | 1100 | 498 | 10 | ||
| E. coli | 5967 | 511(8.6) | 4938 | 1515 | 161 | ||
| Non-Coliform | 60 | 5(8.3) | 35 | 13 | 7 | ||
| Other Coliform | 60 | 4(6.7) | 28 | 13 | 7 | ||
| Streptococcus agalactiae | 1027 | 56(5.5) | 468 | 246 | 30 | ||
| Streptococcus dysgalactiae | 1009 | 28(2.8) | 238 | 108 | 25 | ||
| Streptococcus uberis | 1009 | 22(2.2) | 188 | 83 | 25 | ||
| Streptococcus spp. | 60 | 4(6.7) | 28 | 12 | 7 | ||
| Salmonella spp. | 2791 | 168(6) | 1570 | 548 | 94 | ||
| Listeria spp. | 407 | 85(20.9) | 324 | 32 | 9 | ||
| Enterobacter spp. | 303 | 4(1.3) | 36 | 6 | 9 | ||
| Klebsiella spp. | 303 | 6(2) | 54 | 14 | 9 | ||
| Micrococcus spp. | 321 | 9(2.8) | 55 | 16 | 14 | ||
| Enterococcus spp. | 18 | 1(5.6) | 5 | 0 | 5 | ||
| Proteus spp. | 120 | 9(7.5) | 63 | 12 | 7 | ||
| Shigella spp. | 120 | 21(17.5) | 147 | 25 | 7 | ||
| Food (Meat) | 37 | Salmonella spp. | 2131 | 252(11.8) | 3625 | 1051 | 102 |
| Salmonella Kastrup | 237 | 3(1.3) | 35 | 2 | 8 | ||
| Salmonella Larochelle | 237 | 11(4.6) | 84 | 9 | 6 | ||
| Salmonella enterica subsp.enterica | 150 | 2(1.3) | 70 | 0 | 5 | ||
| non-typhoidal Salmonella | 300 | 17(5.7) | 170 | 61 | 10 | ||
| Salmonella typhimurium | 209 | 2(1) | 36 | 0 | 18 | ||
| Salmonella bovismorbificans | 209 | 4(1.9) | 72 | 0 | 18 | ||
| Salmonella braenderup | 209 | 2(1) | 36 | 15 | 18 | ||
| Salmonella livingstone | 209 | 1(0.5) | 18 | 0 | 18 | ||
| Salmonella hadar | 209 | 1(0.5) | 18 | 13 | 18 | ||
| Salmonella blockley | 209 | 1(0.5) | 18 | 13 | 18 | ||
| Salmonella dublin | 288 | 13(4.5) | 169 | 42 | 26 | ||
| E. coli | 7012 | 389(5.5) | 4188 | 1714 | 250 | ||
| Staphylococus aureus | 1758 | 398(22.6) | 3053 | 1107 | 96 | ||
| Staphylococus spp. | 193 | 92(47.7) | 645 | 258 | 15 | ||
| Staphylococcus intermedius | 181 | 6(3.3) | 165 | 10 | 15 | ||
| Staphylococcus hyicus | 181 | 24(13.3) | 143 | 75 | 13 | ||
| CNS | 181 | 24(13.3) | 165 | 37 | 15 | ||
| Campylobacter jejuni | 1240 | 110(8.9) | 710 | 114 | 26 | ||
| Campylobacter coli | 1240 | 104(8.4) | 178 | 35 | 26 | ||
| Campylobacter lari | 860 | 53(6.2) | 54 | 17 | 21 | ||
| Listeria monocytogenes | 873 | 36(4.1) | 468 | 76 | 13 | ||
| Shigella spp. | 306 | 32(10.5) | 256 | 60 | 8 | ||
| Food (Egg) | 4 | Salmonella spp. | 1127 | 180(16) | 1934 | 853 | 43 |
| Staphylococus aureus | 385 | 96(24.9) | 1056 | 322 | 11 | ||
| E. coli | 385 | 116(30.1) | 1276 | 696 | 11 | ||
| Food (non-animal origin) | 10 | E. coli | 888 | 216(24.3) | 2340 | 607 | 67 |
| Salmonella spp. | 596 | 121(20.3) | 1181 | 328 | 63 | ||
| Staphylococus aureus | 1198 | 181(15.1) | 1818 | 491 | 56 | ||
| Citobacter Spp. | 72 | 9(12.5) | 153 | 11 | 17 | ||
| Enterobacter spp. | 72 | 21(29.2) | 85 | 5 | 17 | ||
| Klebsiella spp. | 572 | 87(15.2) | 999 | 145 | 28 | ||
| Proteus spp. | 72 | 7(9.7) | 85 | 7 | 17 | ||
| Listeria monocytogenes | 384 | 24(6.3) | 240 | 41 | 10 | ||
| Shigella spp. | 540 | 34(6.3) | 350 | 40 | 19 | ||
| Environment | 10 | Acinetobacter spp. | 494 | 13(2.6) | 167 | 87 | 32 |
| Bacillus spp. | 344 | 51(14.8) | 714 | 357 | 14 | ||
| Citobacter Spp. | 839 | 90(10.7) | 666 | 293 | 54 | ||
| Citrobacter diversus | 150 | 9(6) | 99 | 19 | 11 | ||
| CNS | 525 | 110(21) | 1253 | 570 | 27 | ||
| E. coli | 2813 | 427(15.2) | 1871 | 675 | 122 | ||
| Enterobacter aerogenes | 174 | 7(4) | 81 | 28 | 23 | ||
| Enterobacter cloacae | 174 | 24(13.8) | 270 | 111 | 23 | ||
| Enterobacter spp. | 525 | 44(8.4) | 503 | 178 | 23 | ||
| Enterococcus spp. | 344 | 14(4.1) | 196 | 38 | 14 | ||
| Klebsiella oxytoca | 114 | 20(17.5) | 359 | 95 | 31 | ||
| Klebsiella ozaenae | 150 | 4(2.7) | 44 | 18 | 11 | ||
| Klebsiella pneumoniae | 445 | 69(15.5) | 943 | 207 | 56 | ||
| Klebsiella spp. | 525 | 28(5.3) | 301 | 150 | 23 | ||
| Proteus spp. | 525 | 5(1) | 56 | 25 | 23 | ||
| Providencia spp. | 675 | 51(7.6) | 562 | 211 | 34 | ||
| Providencia stuartii | 150 | 8(5.3) | 88 | 29 | 11 | ||
| Pseudomonas aeruginosa | 634 | 41(6.5) | 142 | 94 | 29 | ||
| Pseudomonas mirabilis | 140 | 35(25) | 40 | 21 | 8 | ||
| Salmonella saintpaul | 668 | 28(4.2) | 196 | 26 | 7 | ||
| Salmonella spp. | 1405 | 28(2) | 400 | 139 | 42 | ||
| Serratia spp. | 675 | 20(3) | 232 | 22 | 32 | ||
| Shigella dysenteriae | 150 | 13(8.7) | 143 | 38 | 11 | ||
| Shigella spp. | 1381 | 15(1.1) | 203 | 65 | 30 | ||
| Staphylococcus aureus | 2175 | 144(6.6) | 1773 | 690 | 69 | ||
| Staphylococcus epidermidis | 150 | 5(3.3) | 75 | 20 | 15 | ||
| Staphylococcus saprophyticus | 150 | 2(1.3) | 30 | 6 | 15 | ||
| Streptococcus pyogenes | 150 | 4(2.7) | 214 | 17 | 29 | ||
| Total | 108,930 | 8907 | 86,813 | 4097 |
Table A.2.
Subgroup analysis by bacteria isolated and their pooled prevalence of AMR in different sample type in Ethiopia.
| Sample type | Major bacteria identified and tested | Pooled prevalence of AMR (95% CI) | Heterogeneity |
|||
|---|---|---|---|---|---|---|
| I2% | Q | DF | P-Value | |||
| Food animal | Salmonella kentucky | 53% (37% - 70%) | 64.8% | 173.31 | 61 | 0.00 |
| Pasteurella multocida | 48% (27% -69%) | 82.9% | 76.02 | 13 | 0.00 | |
| Pasteurella haemolytica | 47% (34% - 61) | 90.14% | 446.04 | 44 | 0.00 | |
| Staphylococcus aureus | 37% (20% - 54%) | 90.03% | 381.14 | 38 | 0.00 | |
| E. coli | 37% (24% - 50%) | 95.11% | 920.11 | 45 | 0.00 | |
| Campylobacter jejuni | 30% (18% - 43%) | 98.93% | 4009.29 | 43 | 0.00 | |
| Salmonella Haifa | 30% (17% - 45%) | 8.96% | 52.72 | 48 | 0.30 | |
| Salmonella saintpaul | 22% (12% - 34%) | 79.49% | 292.55 | 60 | 0.00 | |
| Salmonella spp. other than mentioned | 22% (16% - 28%) | 91.39% | 2195.45 | 189 | 0.00 | |
| Salmonella braenderu | 20% (5% - 39%) | 12.32% | 36.49 | 32 | 0.27 | |
| Salmonella mikawasim | 16% (1% - 39%) | 0% | 19.86 | 29 | 0.90 | |
| Salmonella virchow | 15% (7% - 25%) | 54.54% | 92.38 | 42 | 0.00 | |
| Salmonella Dublin | 12% (1% - 29%) | 84.74% | 190.08 | 29 | 0.00 | |
| Salmonella typhimurium | 7% (3% - 13%) | 33.59% | 93.35 | 62 | 0.01 | |
| Campylobacter coli | 7% (1% - 18%) | 95.4% | 565.18 | 26 | 0.00 | |
| Overall | 20% (16% - 24%) | 90.73% | 11,207 | 1039 | 0.00 | |
| Milk and milk product | Staphylococcus spp. other than Staphylococcus aureus | 70% (56% - 82%) | 82.59% | 229.78 | 40 | 0.00 |
| Streptococcus agalactiae | 53% (31% - 75%) | 91.07% | 246.49 | 22 | 0.00 | |
| Streptococcus uberis | 46% (28% - 64%) | 81.3% | 128.37 | 24 | 0.00 | |
| CNS | 40% (21% - 60%) | 94.98% | 438.5 | 22 | 0.00 | |
| Streptococcus dysgalactiae | 38% (25% - 51%) | 81.26% | 165.43 | 31 | 0.00 | |
| Salmonella spp. | 29% (21% - 38%) | 89.95% | 925.14 | 93 | 0.00 | |
| E. coli | 28% (22% - 33%) | 93.94% | 2639.6 | 160 | 0.00 | |
| Staphylococcus aureus | 27% (23% - 31%) | 97.38% | 14,424.6 | 378 | 0.00 | |
| Micrococcus spp. | 24% (10% - 39%) | 10.1% | 14.46 | 13 | 0.34 | |
| Staphylococcus epidermidis | 18% (8% - 30%) | 85.44% | 233.48 | 34 | 0.00 | |
| Staphylococcus intermedius | 5% (0% - 26%) | 61.35% | 38.8 | 15 | 0.00 | |
| Overall | 29% (26% - 32%) | 95.55% | 20,925.7 | 932 | 0.00 | |
| Meat | E. coli | 41% (33% - 48%) | 94.54% | 4564.25 | 249 | 0.00 |
| Staphylococcus aureus | 30% (19% - 42%) | 96.79% | 2961.19 | 95 | 0.00 | |
| Salmonella spp. | 28% (20% - 36%) | 95.5% | 2247.11 | 101 | 0.00 | |
| Campylobacter lari | 24% (5% - 48%) | 56.39% | 45.87 | 20 | 0.00 | |
| Campylobacter coli | 17% (9% - 26%) | 43.46% | 44.21 | 25 | 0.01 | |
| Campylobacter jejuni | 13% (5% - 23%) | 91.83% | 305.99 | 25 | 0.00 | |
| Overall | 28% (24% - 32%) | 93.64% | 11,990.2 | 762 | 0.00 | |
| FNA | Staphylococcus aureus | 20% (10% - 32%) | 96.51% | 1575.16 | 55 | 0.00 |
| E. coli | 20% (12% - 30%) | 96.22% | 1747.65 | 66 | 0.00 | |
| Salmonella spp. | 16% (7% - 27%) | 94.31% | 1088.9 | 62 | 0.00 | |
| Shigella spp. | 11% (2% - 24) | 88.62% | 158,21 | 18 | 0.00 | |
| Klebsiella spp. | 5% (0% - 13%) | 91.5% | 317.79 | 27 | 0.00 | |
| Overall | 13% (10% - 18%) | 94.41% | 5244.49 | 293 | 0.00 | |
| Food handlers | Salmonella spp. | 37% (29% - 46%) | 77.88% | 298.42 | 66 | 0.00 |
| E. coli | 36% (22% - 51%) | 80.35% | 91.59 | 18 | 0.00 | |
| Shigella spp. | 35% (23% - 48%) | 83.01% | 200.17 | 34 | 0.00 | |
| Staphylococcus aureus | 16% (6% - 29%) | 96.89% | 706.86 | 22 | 0.00 | |
| Overall | 29% (23% - 35%) | 88.46% | 1576.57 | 182 | 0.00 | |
| Environment | Pseudomonas aeruginosa | 67% (47% - 84%) | 73.41% | 105.29 | 28 | 0.00 |
| Citobacter spp. | 47% (32% - 61%) | 90.75% | 573.21 | 53 | 0.00 | |
| Acinetobacter spp. | 44% (25% - 63) | 79.13% | 148.57 | 31 | 0.00 | |
| Proteus spp. | 39% (16% - 63) | 55.39% | 49.32 | 22 | 0.00 | |
| Enterobacter spp. | 31% (17% - 46%) | 92.02% | 275.72 | 22 | 0.00 | |
| Staphylococcus aureus | 30% (20% - 40%) | 94.94% | 1342.58 | 68 | 0.00 | |
| E. coli | 29% (21% - 37%) | 90.28% | 1244.39 | 121 | 0.00 | |
| Salmonella spp. | 28% (16% - 43%) | 87.06% | 316.93 | 41 | 0.00 | |
| Shigella spp. | 28% (11% - 46%) | 78.29% | 133.61 | 29 | 0.00 | |
| Providencia spp. | 28% (15% - 43%) | 91% | 366.8 | 33 | 0.00 | |
| Klebsiella pneumonia | 20% (13% - 27%) | 83.75% | 326.22 | 53 | 0.00 | |
| Serratia spp. | 5% (1% - 11%) | 51.43% | 67.94 | 33 | 0.00 | |
| Overall | 29% (25% - 32%) | 89.52% | 7807.47 | 818 | 0.00 | |
References
- 1.Queenan K., Häsler B., Rushton J. A one health approach to antimicrobial resistance surveillance: is there a business case for it? Int. J. Antimicrob. Agents. 2016;48:422–427. doi: 10.1016/j.ijantimicag.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 2.Grace D. Nirobi, Kenya; International Livestock Research Institute: 2015. Review of Evidence on Antimicrobial Resistance and Animal Agriculture in Developing Countries. [Google Scholar]
- 3.Jasovsky D., Littmann J., Zorzet A., Cars O. Antimicrobial resistance - a threat to the world’s sustainable development. Antimicrob. Resist. 2016 doi: 10.1159/isbn.978-3-8055-9324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang K.L., Caffrey N.P., Nóbrega D.B., Cork S.C., Ronksley P.E., Barkema H.W. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planet. Health. 2017;1:316–327. doi: 10.1016/S2542-5196(17)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Julian D. Antibiotic resistance in and from nature. One Heal. 2014;1:185–194. doi: 10.1128/microbiolspec.oh-0005-2012. American Society of Microbiology. [DOI] [Google Scholar]
- 6.Butaye P., Duijkeren E., Prescott J.F., Schwarz S. Antimicrobial resistance in bacteria from animals and the environment. Vet. Microbiol. 2014:269–272. doi: 10.1016/j.vetmic.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Prescott J.F. The resistance tsunami, antimicrobial stewardship, and the golden age of microbiology. Vet. Microbiol. 2014:273–278. doi: 10.1016/j.vetmic.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 8.Woolhouse M., Ward M., Van Bunnik B., Farrar J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R Soc. B Biol. Sci. 2015 doi: 10.1098/rstb.2014.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathew A.G., Cissell R., Liamthong S. Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog. Dis. 2007 doi: 10.1089/fpd.2006.0066. [DOI] [PubMed] [Google Scholar]
- 10.Heuer H., Schmitt H., Smalla K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011 doi: 10.1016/j.mib.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Elton L., Thomason M.J., Tembo J., Velavan T.P., Pallerla S.R., Arruda L.B. Antimicrobial resistance preparedness in sub-Saharan African countries. Antimicrob. Resist. Infect. Control. 2020 doi: 10.1186/s13756-020-00800-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization . 2020. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report. [Google Scholar]
- 13.WHO 2020. https://apps.who.int/iris/bitstream/handle/10665/332081/9789240005587-eng.pdf?ua=1
- 14.Seale A.C., Hutchison C., Fernandes S., Stoesser N., Kelly H., Lowe B. Supporting surveillance capacity for antimicrobial resistance: laboratory capacity strengthening for drug resistant infections in low and middle income countries. Wellcome Open Res. 2017 doi: 10.12688/wellcomeopenres.12523.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seale A.C., Gordon N.C., Islam J., Peacock S.J., Scott J.A.G. AMR surveillance in low and middle-income settings - a roadmap for participation in the Global Antimicrobial Surveillance System (GLASS) Wellcome Open Res. 2017 doi: 10.12688/wellcomeopenres.12527.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibrahim R.A., Teshale A.M., Dinku S.F., Abera N.A., Negeri A.A., Desta F.G. Antimicrobial resistance surveillance in Ethiopia: implementation experiences and lessons learned. Afr. J. Lab. Med. 2018;8:1–4. doi: 10.4102/ajlm.v7i2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moges F., Endris M., Mulu A., Tessema B., Belyhun Y., Shiferaw Y. The growing challenges of antibacterial drug resistance in Ethiopia. J. Glob. Antimicrob. Resist. 2014;2:148–154. doi: 10.1016/j.jgar.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Seboxa T., Amogne W., Abebe W. High mortality from blood stream infection in Addis Ababa, Ethiopia, is due to antimicrobial resistance. PLoS One. 2015;10 doi: 10.1371/journal.pone.0144944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abebe W., Tinsae T., Kong L., Temesgen A., Barbara J., Alina D. Alarming rates of drug-resistance in gram-negative blood stream infections among patients in Ethiopia (P0947) 28th Eur. Congr. Clin. Microbiol. Infect. Dis. 2018 Madrid. [Google Scholar]
- 20.Moher D., Shamseer L., Clarke M., Ghersi D., Liberati A., Petticrew M. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015;4:1–9. doi: 10.1186/2046-4053-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DerSimonian R., Laird N. Meta-analysis in clinical trials. Control. Clin. Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 22.Higgins J.P.T., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 23.Vernet Guy, Mary Catherine, Altmann Dany M., Doumbo Ogobara, Morpeth Susan, Bhutta Zulfiqar A., KPK Surveillance for antimicrobial drug resistance in under-resourced countries. Emerg. Infect. Dis. 2014;20:434–441. doi: 10.3201/eid2003.121157. https://doi.org/doi: 10.3201/eid2003.121157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayukekbong J.A., Ntemgwa M., Atabe A.N. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob. Resist. Infect. Control. 2017;6 doi: 10.1186/s13756-017-0208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gemeda B.A., Amenu K., Magnusson U., Dohoo I., Hallenberg G.S., Alemayehu G. Antimicrobial use in extensive smallholder livestock farming systems in Ethiopia: knowledge, attitudes, and practices of livestock keepers. Front. Vet. Sci. 2020;7 doi: 10.3389/fvets.2020.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zelalem A., Sisay M., Vipham J.L., Abegaz K., Kebede A., Terefe Y. The prevalence and antimicrobial resistance profiles of bacterial isolates from meat and meat products in Ethiopia: a systematic review and meta-analysis. Int. J. Food Contam. 2019;6 doi: 10.1186/s40550-019-0071-z. [DOI] [Google Scholar]
- 27.Mouiche M.M.M., Moffo F., Akoachere J.F.T.K., Okah-Nnane N.H., Mapiefou N.P., Ndze V.N. Antimicrobial resistance from a one health perspective in Cameroon: a systematic review and meta-analysis. BMC Public Health. 2019;19:1–20. doi: 10.1186/s12889-019-7450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moges F., Endris M., Mulu A., Tessema B., Belyhun Y., Shiferaw Y. The growing challenges of antibacterial drug resistance in Ethiopia. J. Glob. Antimicrob. Resist. 2014;2:148–154. doi: 10.1016/j.jgar.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Hu Y., Gao G.F., Zhu B. The antibiotic resistome: gene flow in environments, animals and human beings. Front. Med. 2017 doi: 10.1007/s11684-017-0531-x. [DOI] [PubMed] [Google Scholar]
- 30.White A., Hughes J.M. Critical importance of a one health approach to antimicrobial resistance. Ecohealth. 2019 doi: 10.1007/s10393-019-01415-5. [DOI] [PubMed] [Google Scholar]
- 31.Dolejska M., Literak I. Wildlife is overlooked in the epidemiology of medically important antibiotic-resistant bacteria. Antimicrob. Agents Chemother. 2019 doi: 10.1128/AAC.01167-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sisay M., Worku T., Edessa D. Microbial epidemiology and antimicrobial resistance patterns of wound infection in Ethiopia: a meta-analysis of laboratorybased cross-sectional studies. BMC Pharmacol. Toxicol. 2019 doi: 10.1186/s40360-019-0315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muhie O.A. Antibiotic use and resistance pattern in Ethiopia: systematic review and meta-analysis 2019. Arti. 2019;8 doi: 10.1155/2019/2489063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mezali L., Hamdi T.M. Prevalence and antimicrobial resistance of Salmonella isolated from meat and meat products in Algiers (Algeria) Foodborne Pathog. Dis. 2012;9:522–529. doi: 10.1089/fpd.2011.1032. [DOI] [PubMed] [Google Scholar]
- 35.Kikuvi G.M., Ombui J.N., Mitema E.S. Serotypes and antimicrobial resistance profiles of Salmonella isolates from pigs at slaughter in Kenya. J. Infect. Dev. Ctries. 2010;4:243–248. doi: 10.3855/jidc.446. [DOI] [PubMed] [Google Scholar]
- 36.Fortini D., Fashae K., García-Fernández A., Villa L., Carattoli A. Plasmidmediated quinolone resistance and β-lactamases in Escherichia coli from healthy animals from Nigeria. J. Antimicrob. Chemother. 2011;66:1269–1272. doi: 10.1093/jac/dkr085. [DOI] [PubMed] [Google Scholar]