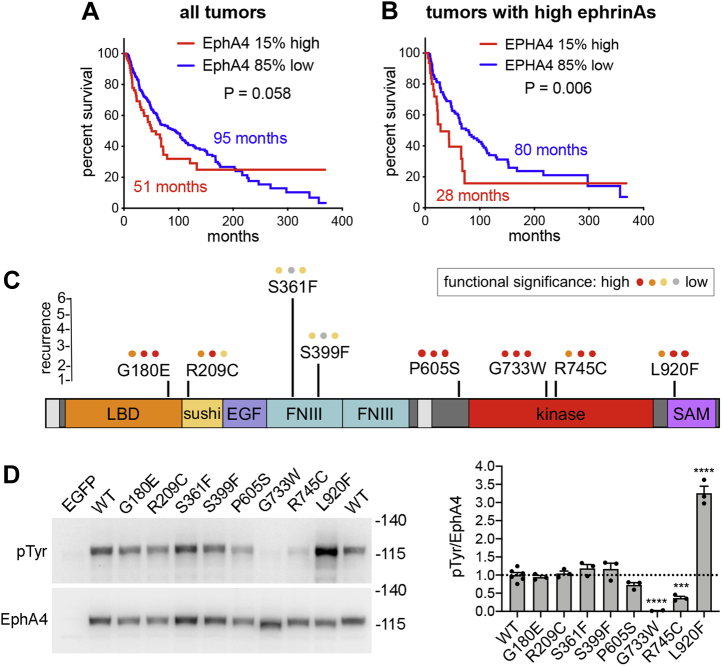
Figure 1.
Correlation of EphA4 expression with patient survival and EphA4 mutations in melanoma.A, the correlation between high (top 15%) EphA4 mRNA expression and decreased overall patient survival does not reach statistical significance when considering all melanoma tumors. B, high EphA4 expression in the subset of tumors with highest (top 15%) mRNA expression of one or more of the five ephrinA ligands significantly correlates with decreased patient survival. In both A and B, the 15% of tumor samples with highest EphA4 expression were compared to the 85% remaining tumor samples. B includes for each ephrinA ligand the 15% of tumors with highest expression, for a total of ∼45% of all the tumors. mRNA expression z-scores relative to all samples (log RNA Seq V2 RSEM) from the TCGA Firehose Legacy skin cutaneous melanoma dataset (n = 472 tumor samples with mRNA expression data) were used for analysis. Median survival times are indicated in the graphs and p values were calculated using the log-rank Mantel–Cox test. C, the location of the eight EphA4 melanoma mutations analyzed is shown in relation to the EphA4 domain structure. The mutations were selected for further investigation from 12 skin melanoma studies available in the cBioPortal website (cbioportal.org). The height of the black vertical lines indicates the number of tumors with that particular mutation. The colored dots above the name of each mutation indicate the prediction of functional significance according to three prediction programs: Mutation Assessor, SIFT and PolyPhen-2 (cbioportal.org). The EphA4 signal peptide and transmembrane helix are shown in light gray and linkers are shown in dark gray, including the juxtamembrane segment containing the P605S mutation. D, EphA4 WT, the eight EphA4 mutants, and EGFP as a control were transiently expressed in HEK293 cells and cell lysates were probed by immunoblotting with antibodies to phosphotyrosine (pTyr) and to EphA4. The bar graph shows averages and standard errors from quantifications of three experiments (individual values from each experiment are shown as dots). ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001 for the comparison with WT by one-way ANOVA. EGF, epidermal growth factor-like domain; FNIII, fibronectin type III domain; LBD, ligand-binding domain; kinase, kinase domain; SAM, sterile alpha motif domain; sushi, sushi domain.