Figure 3.
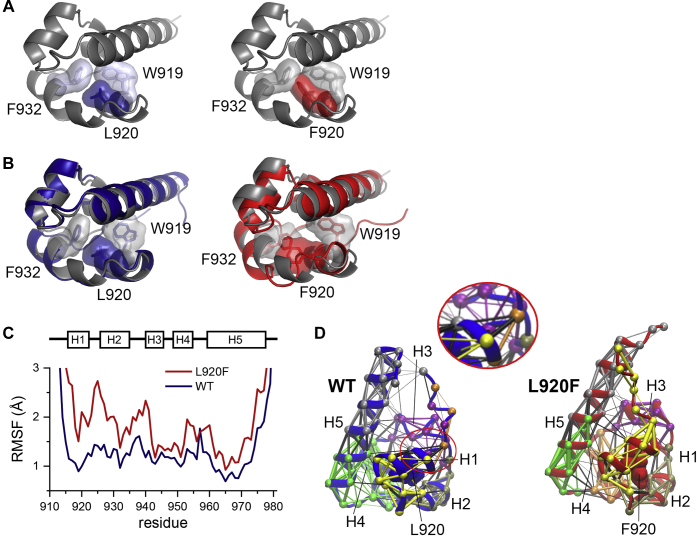
All-atom MD simulations suggest that the EphA4 L920F mutation introduces local and global structural perturbations.A, structure of the EphA4 WT SAM domain. (left; PDB ID: 1B0X; (30)) and model of the L920F mutant (right), obtained by direct substitution of L920 with phenylalanine, in ribbon representation with the indicated residues shown as sticks and as a molecular surface. The model illustrates how W919 and F932 clash with a phenylalanine at position 920. B, representative structures obtained from MD simulations of the EphA4 WT SAM domain (left) and EphA4 L920F SAM domain (right) are aligned with the EphA4 SAM domain crystal structure (gray) by minimizing backbone RMSD values. C, the RMSF values calculated using the crystal structure coordinates as the reference are plotted for each residue in the EphA4 WT and L920F SAM domains over the course of the MD simulations, each averaged over three replicates. A schematic of the SAM domain helix positions is shown above the figure. D, dynamic network analysis for the EphA4 WT and L920F mutant SAM domain structures. Nodes, indicated by spheres, highlight the α-carbon atoms and the thickness of the edges (lines connecting the nodes) is proportional to the correlation of atomic motion in space and time. The WT L920 or mutant F920 is rendered with atoms as spheres. Six communities (sets of residues that exhibit coordinated motion) are shown in different colors: community 1 in yellow, community 2 in olive, community 3 in purple, community 4 in orange, community 5 in green, and community 6 in gray. Edges drawn within the same community are colored according to that community and edges drawn between nodes of different communities are colored black. The α-helices (H1 through H5) are indicated and the portion of H1 circled in red in the WT SAM domain is also shown as an enlargement to highlight the different communities in this α-helix.