Figure 6.
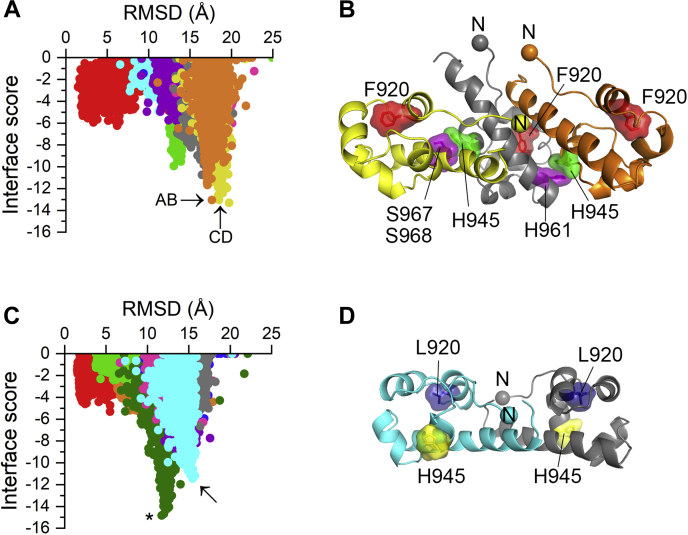
In silico docking predicts two stable interfaces for the EphA4 L920F SAM domain.A, ten EphA4 L920F SAM dimer structures generated by ClusPro were optimized by creating 2000 decoys for each with PyRosetta. Interface scores for the decoys are plotted as a function of the RMSD value calculated relative to the initial ClusPro structure “0” (ClusPro outputs are numbered 0–9). Each of the ten sets of decoys is shown in a different color. The lowest-energy decoys from each set represent the optimized dimer structures. Two lowest-energy dimer structures for the EphA4 L920F SAM domain, referred to as AB (orange) and CD (yellow), are indicated by arrows. The decoy with the lowest interface score for the set of structures colored in yellow was considered an outlier (see Fig. S5), so the second lowest was selected. B, model of an EphA4 L920F SAM domain trimer that engages both AB and CD interfaces. The three SAM domains in the trimer are shown in orange (molecule A), yellow (molecule D), and gray (molecule B/C). Residue F920 is shown in red as sticks and as a molecular surface. Residue H945, shown in green as sticks and as a molecular surface, stabilizes both interfaces (see Table S1) by engaging H961 (purple sticks and molecular surface) in molecule B/C and S967/S968 (purple sticks and molecular surface) in molecule D. C, ten sets of EphA4 WT decoys were generated as described in A. A single lowest-energy structure, indicated by an asterisk, was identified for the EphA4 WT SAM domain (green). However, this model did not match some experimental data and therefore the second lowest structure (cyan), indicated by an arrow, was selected for further analysis. D, structural representation of the second lowest-energy EphA4 WT dimer structure in which the two SAM domains (A and B) are indicated in cyan and gray. Residue L920 is indicated in blue and residue H945 in yellow, both shown as sticks and as a molecular surface. This model is structurally similar to the crystallographic dimer shown in Fig. S6C.