Abstract
Death receptor 4 (DR4) is a cell surface protein that is generally thought to mediate apoptosis upon binding to its ligand named TRAIL. However, its contribution to apoptosis resistance has also been reported. MET (or c-MET) gene amplification represents an important mechanism for acquired resistance to EGFR tyrosine kinase inhibitors (EGFR-TKIs) against EGFR mutant non-small cell lung cancer (NSCLC). This study focuses on demonstrating the impact of MET inhibition on DR4 modulation in MET-amplified EGFR mutant NSCLC cell lines and the underlying mechanisms. Several MET inhibitors decreased DR4 levels in MET-amplified HCC827 cell lines resistant to EGFR-TKIs with no or limited effects on modulating DR5 levels, while increasing DR4 levels in HCC827 parental cells and other NSCLC cell lines. MET inhibitors did not affect DR4 stability, but decreased DR4 mRNA levels with suppression of AP-1-dependent DR4 promoter transactivation. Moreover, these inhibitors suppressed ERK and c-Jun phosphorylation accompanied with decreasing c-Jun levels. Hence, it is likely that MET inhibition downregulates DR4 expression in MET-amplified EGFR mutant NSCLC cells through suppressing AP-1-mediated DR4 transcription. Osimertinib combined with MET inhibition synergistically induces apoptosis in the MET-amplified EGFR mutant NSCLC cells accompanied with augmented DR4 reduction both in vitro and in vivo. Furthermore, MET inhibition combined with TRAIL enhanced killing of MET-amplified EGFR mutant HCC827/AR cells, but not HCC827 parental cells. These data collectively suggest that DR4 may possess an unrecognized anti-apoptotic function, contributing to apoptosis resistance under given conditions.
Keywords: MET, DR4, EGFR-TKIs, Acquired resistance, Lung cancer
Abbreviations: DR4, death receptor4; DR5, death receptor 5; EGFR, epidermal growth factor recetptor; EGFR-TKI, EGFR tyrosine kinase inhibitor; NSCLC, non-small cell lung cancer; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; Act D, actinomycin D; CHX, cycloheximide; MFI, mean fluorescent intensity; SRB, sulforhodamine B; siRNA, small intergering RNA
Introduction
MET (or c-MET) gene amplification, which is detectable in approximately 5%-22% of resistant non-small cell lung cancer (NSCLC) tumors harboring activating EGFR mutations (e.g., 19del and L858R), represents an important resistance mechanism to the first generation EGFR tyrosine kinase inhibitors (EGFR-TKIs) in addition to acquisition of a secondary T790M mutation that accounts for approximately 60% of resistant cases [1], [2], [3]. Moreover, MET gene amplification was also detected in the clinic from EGFR mutant (EGFRm) NSCLC patients receiving third generation EGFR-TKIs and from osimertinib-resistant NSCLC cells in the lab by us [4] and others [5], [6], [7]. Our previous work has clearly demonstrated that MET amplification and protein hyperactivation confers resistance to third generation EGFR-TKIs as well [4].
Death receptor 4 (DR4), also known as tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) receptor 1 (TRAIL-R1) or tumor necrosis factor receptor superfamily member 10A (TNFRSF10A), is a cell surface protein for TRAIL. Similar to its sibling, death receptor 5 (DR5), DR4, when ligated with TRAIL, induces apoptosis through the specific interaction of trimerized DR4 with Fas-associated death domain (FADD) and the subsequent recruitment of caspase-8 and ultimate caspase-8-dependent apoptosis [8,9]. It is known that the main effector immune cell types exerting cytotoxicity against malignant cells are cytotoxic T lymophocytes (CTLs) and natural killer (NK) cells, which can generate and secrete TRAIL. Thus, the induction of apoptosis by ligation of endogenous TRAIL with its receptors on cancer cells has also been recognized as a critical mechanism accounting for immune surveillance against malignant cells [10], [11], [12].
DR4 generally shares a redundant function with DR5 in mediating TRAIL-induced apoptosis as discussed above. However, DR4 does display some distinct functions from DR5. For example, while DR5 knockdown protected cancer cells from undergoing TRAIL-induced apoptosis, DR4 knockdown substantially enhanced TRAIL-induced apoptosis [13]. Interestingly, both DR4 and DR5 expression are positively regulated by MEK/ERK signaling despite through differential transcriptional mechanisms [14], [15], [16].
Since MET activation activates the MEK/ERK signaling pathway, we compared the effects of MET inhibitors on the growth of MET-amplified EGFR-TKI resistant NSCLC cells, modulation of MEK/ERK signaling and regulation of the expression of DR4 and DR5 in these cell lines with their counterpart parental cells. We found that these MET-amplified EGFR-TKI resistant NSCLC cell lines responded better than their parental cells to different MET inhibitors with the suppression of MEK/ERK signaling and downregulation of DR4. Hence, this study focused on studying the underlying mechanism accounting for DR4 downregulation and the biological significance of DR4 suppression.
Materials and methods
Reagents
The resources and preparation of osimertinib (AZD9291), crizotinib (PF02341066), SGX523, ARQ197, MG132, actinomycin D (Act D) and cycloheximide (CHX) were the same as described previously [4,17]. Human recombinant TRAIL was purchased from PeproTech, Inc. (Rocky Hill, NJ). Mouse (B-N28) and rabbit (D9S1R) monoclonal DR4 antibodies were purchased from Cell Science (Newburyport MA) and Cell Signaling Technology, Inc. (Beverly, MA), respectively. Rabbit monoclonal DR5 antibody (D4E9) was purchased from Cell Signaling Technology, Inc. Other antibodies were the same as described in our previous studies [4,14,16,17].
Cell lines and cell culture
HCC827, HCC827/ER (erlotinib-resistant), HCC827/AR (AZD9291-resistant) and other NSCLC cell lines and their culture conditions were the same as described previously [4,17].
Cell survival assay
Cells seeded in 96-well plates at the appropriate densities for overnight were exposed to the tested drugs. After 3 d, cells numbers were measured by sulforhodamine B (SRB) assay as previously described [18].
Cell surface DR4 detection
Cell surface DR4 expression was detected with flow cytometry as described previously [19]. The mean fluorescent intensity (MFI) that represents antigenic density on a per cell basis was used to assess cell surface DR4 levels. Alexa Fluor® 488-conjugated mouse anti-human DR4 (CD261) antibody (DR-4-02) was purchased from Bio-Rad Laboratories (Hercules, CA).
Western blot analysis
Preparation of whole-cell protein lysates and Western blot analysis were described previously [4,17].
Gene knockdown using small interfering RNA (siRNA)
The control and MET siRNAs and transfections of these siRNAs were the same as reported previously [4].
RT-PCR detection of DR4 mRNA
DR4 mRNA was detected with RT-PCR as described previously [20].
Reporter plasmids and luciferase assay
All DR4 reporter constructs used in this study and luciferase assay were the same as described previously [21].
Xenograft tissues
HCC827/AR xenograft tissues receiving vehicle, osimertinib, crizotinib and osimertinib combined with crizotinib were from the same experiment conducted previously [4]. DR4 in these tissues was detected with Western blotting.
Colony formation assay
The procedure for this assay was the same as described previously [4] and used for evaluating the long-term effects of crizotinib combined with TRAIL on cell-killing of the tested NSCLC lines.
Results
MET-amplified EGFRm NSCLC cell lines with acquired resistance to EGFR-TKIs respond better than their parental cells to MET inhibitors
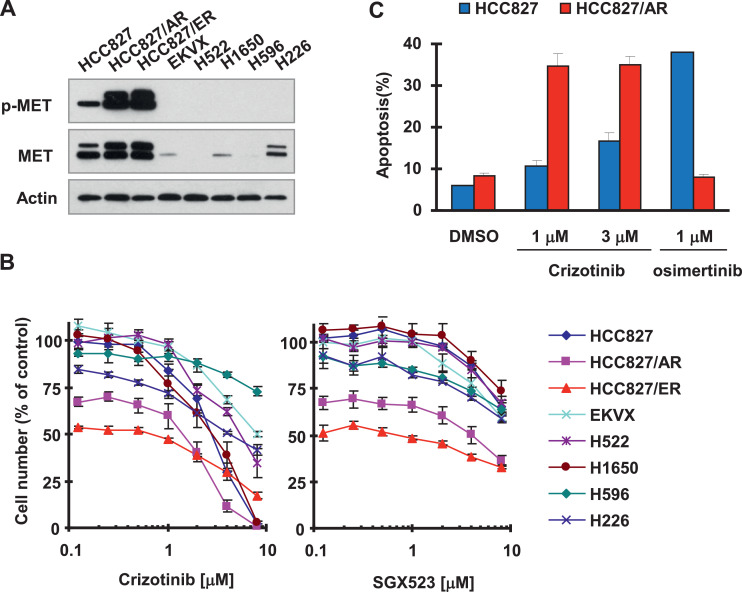
We first compared the effects of MET inhibitors on the growth of MET-amplified NSCLC cell lines (HCC827/ER and HCC827/AR) due to acquired resistance to EGFR-TKIs with its parental HCC827 and other NSCLC cell lines without MET amplification. As validated with Western blotting, both HCC827/ER and HCC827/AR have very high levels of MET and p-MET, whereas other tested cell lines had very low or undetectable levels of these proteins (Fig. 1A). Consistently, these 2 cell lines were sensitive to both crizotinib and SGX523 while other NSCLC cell lines responded poorly to these agents (Fig. 1B). Moreover, HCC827/AR cells were much more sensitive than HCC827 cells to undergoing apoptosis upon crizotinib treatment, evidenced by detection of much more apoptotic cells in HCC827/AR cells than in HCC827 cells (Fig. 1C). As a positive control, we detected a significant increase of apoptotic cells in HCC827 cells exposed to osimertinib, but not in HCC827/AR cells treated with osimertinib (Fig. 1C). Hence the MET-amplified cell lines clearly display increased sensitivity to MET inhibition.
Fig. 1.
Osimertinib- and erlotinib-resistant HCC827 cells display elevated levels of MET and p-MET (A) and are more sensitive to MET inhibitors than their parental and other human NSCLC cell lines (B) including induction of apoptosis (C). A, Whole-cell protein lysates were prepared from different untreated cell lines as indicated for detection of the indicated proteins with Western blotting. B, The panel of human NSCLC cell lines as indicated was seeded in 96-well plates and then treated with different concentrations of crizotinib or SGX523 as indicated on the second day. After 3 d, cell numbers were estimated using the SRB assay. The data are means ± SDs of 4 replicate determinations. C, Both HCC827 and HCC827/AR cell lines were exposed to the indicated treatments. After 48 h, the cells were harvested for detection of apoptosis with annexin V-flow cytometry. Each column represents the mean ± SD of duplicate determination.
MET inhibitors suppress MEK/ERK signaling and decrease DR4 levels with minimal effects on DR5 in MET-amplified EGFRm NSCLC cells
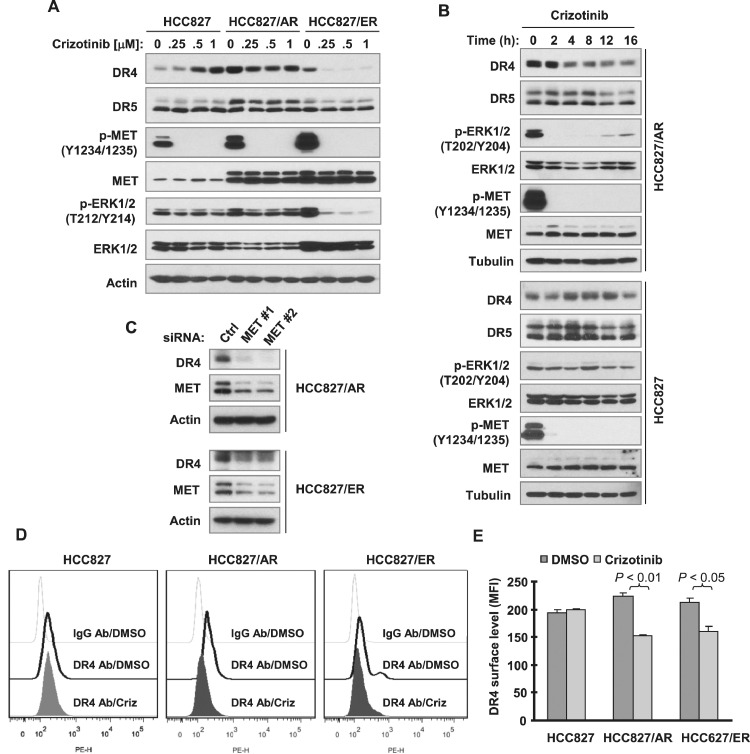
We then compared the effects of crizotinib on the modulation of MEK/ERK signaling and DR4 and DR5 expression between HCC827 and its derived MET-amplified HCC817/ER and HCC827/AR cells. Although this MET inhibitor suppressed MET phosphorylation across the tested cell lines, it inhibited ERK phosphorylation only in HCC827/ER and HCC827/AR cells. Moreover, we detected reduced levels of DR4 in these 2 cell lines. In contrast, we observed that DR4 levels were even increased in HCC827 cells exposed to crizotinib. Interestingly, crizotinib minimally modulated DR5 expression in these cell lines (Figs. 2A and 2B). Moreover, we found that MET inhibition by directly knocking down MET in both HCC827/ER and HCC827/AR decreased DR4 levels (Fig. 2C). In agreement, crizotinib significantly decreased cell surface DR4 levels in both HCC827/ER and HCC827/AR, but not in HCC827 cells (Fig. 2D).
Fig. 2.
Both pharmacological and genetic inhibition of MET decrease DR4 levels (A-C) including cell surface DR4 (D and E) in EGFR-TKI resistant HCC827 cells. A and B, The given cancer cell lines were treated with different concentrations of crizotinib for 8 h (A) or with 200 nM crizotinib for different times as indicated (B). C, Both HCC827/AR and HCC827/ER cells in 6-well plates were transfected with the indicated siRNAs for 48 h. After the aforementioned treatments, the cells were harvested for preparation of whole-cell protein lysates and subsequent Western blotting analysis. D and E, The indicated cell lines were treated with 0.5 μM crizotinib for 12 h and then harvested for detection of cell surface DR4 with flow cytometry. The representative results are shown in D and average data (MFIs) from triplicate assays are presented in E as means ± SDs. The gray dot line open peaks in D represent DMSO-treated cells stained with a matched control PE-conjugated IgG isotype antibody. The black solid line open peaks show DMSO-treated cells stained with PE-conjugated anti-DR4 antibody. The filled peaks represent crizotinib (Criz)-treated cells stained with PE-conjugated anti-DR4 antibody.
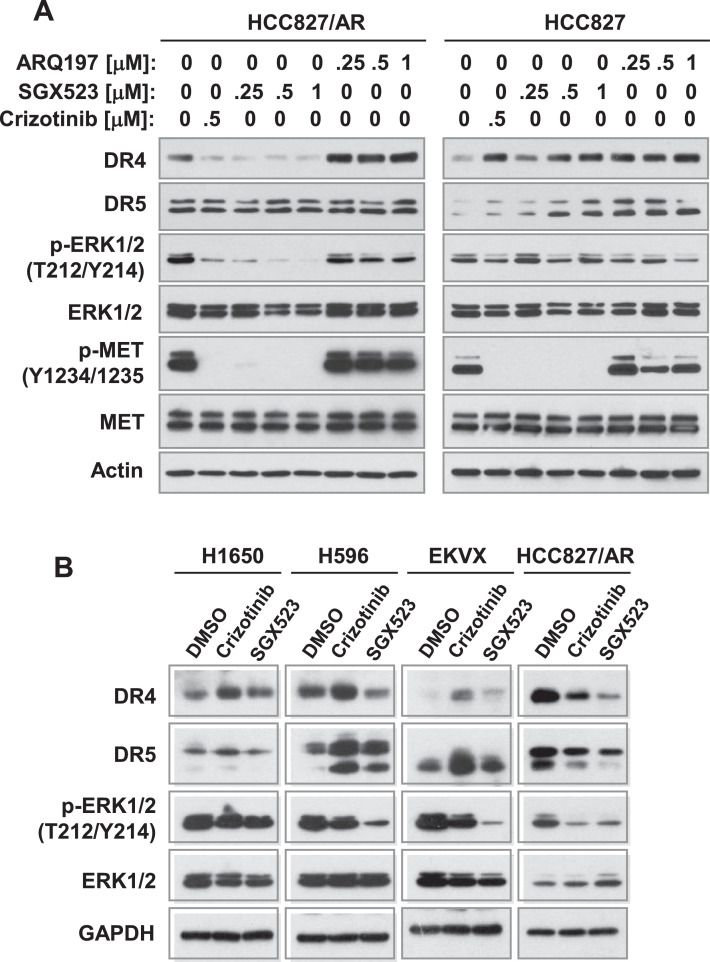
The other MET inhibitor, SGX523, functioned in a similar way to suppress the phosphorylation of MET and ERK and to decrease DR4 levels without affecting DR5 levels in HCC827/AR cells, while it increased both DR4 and DR5 levels without suppressing ERK phosphorylation in HCC827 cells. Although ARQ197 is a MET inhibitor, we found that it did not inhibit phosphorylation of MET and ERK in both HCC827 and HCC827/AR cells, but increased DR4 levels in these cell lines (Fig. 3A). In other NSCLC cell lines without mutant EGFR including H1650, H596 and EKVX, we found that crizotinib increased DR4 levels with limited suppression effects on ERK phosphorylation. SGX523 slightly increased DR4 levels with limited effect on inhibiting ERK phosphorylation in H1650 cells, but decreased DR4 levels in H596 cells with suppression of ERK phosphorylation. Both inhibitors increased DR5 levels in H596 and EKVX cells (Fig. 3B). Taking these findings together, it is clear that MET inhibition, particularly with crizotinib and SGX523, suppresses MEK/ERK signaling and decreases DR4 levels with minimal effects on DR5 in MET-amplified EGFRm NSCLC cells.
Fig. 3.
Comparison of the effects of different MET inhibitors on modulating DR4 levels between MET-amplified EGFR-TKI resistant HCC827 cells and other NSCLC cell lines. The indicated cell lines were exposed to different concentrations of MET inhibitors as indicated (A) or treated with 1 μM of the given MET inhibitors (B) for 8 h. The cells were then harvested for preparation of whole-cell protein lysates and subsequent Western blot analysis for detection of the indicated proteins.
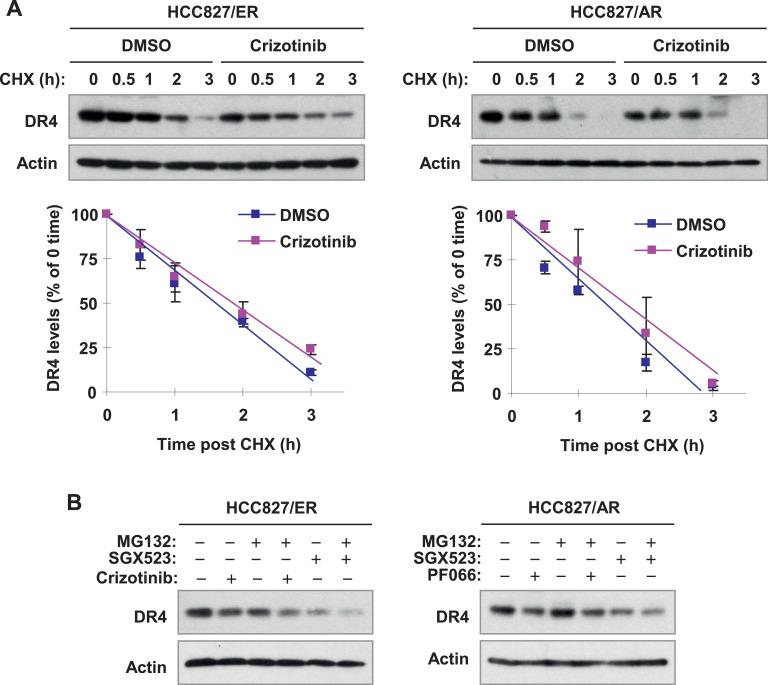
MET inhibitors do not alter DR4 protein stability
To understand the mechanism(s) by which MET inhibitors decrease DR4 levels in MET-amplified cells, we first determined whether MET inhibitors enhances DR4 degradation by conducting a CHX chase assay. In both HCC827/ER and HCC827/AR cells, DR4 degradation rates in cells exposed to DMSO and to crizotinib were comparable (Fig. 4A), indicating that crizotinib does not enhance DR4 degradation or reduce stability. In agreement, the presence of MG132, a widely used proteasome inhibitor, did not prevent DR4 reduction induced by either crizotinib or SGX523 (Fig. 4B). Thus, it is clear that these MET inhibitors do not work at the posttranslational level to decrease DR4 levels in MET-amplified cells.
Fig. 4.
Crizotinib does not alter protein stability (A) and degradation (B) of DR4 in EGFR-TKI resistant HCC827 cells. A, The indicated cells were exposed to 0.5 μM crizotinib for 4 h followed by the addition of 10 μg/ml of CHX. At the indicated times post CHX, the cells were harvested for preparation of whole-cell protein lysates and subsequent Western blot analysis. Protein levels were quantified with NIH ImageJ software, normalized to actin and plotted as percentages of 0 time. The data are means +/- SEs of duplecated assays. B, The indicated cell lines were pre-treated with 10 μg/ml MG132 for 30 min and then co-treated with 0.5 μM MET inhibitors as indicated for an additional 5 h. The cells were then harvested for preparation of whole-cell protein lysates and subsequent Western blot analysis.
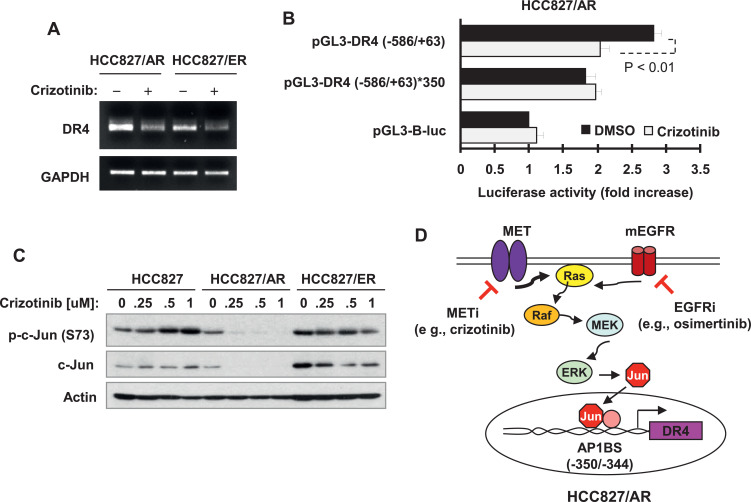
MET inhibitors suppress AP-1-dependent DR4 transcription
We next determined whether crizotinib decreased DR4 mRNA levels. Using RT-PCR, we detected reduced levels of DR4 mRNA in both HCC827/AR and HCC827/ER cells exposed to crizotinib in comparison with DMSO-treated cells (Fig. 5A). Moreover, we found that crizotinib significantly inhibited DR4 promoter activity in the promoter luciferase assay, but lost this activity when the AP-1 site in DR4 promoter region was mutated (Fig. 5B), suggesting that crizotinib suppressed AP-1-dependent DR4 transcription. Furthermore, we examined the effects of crizotinib on c-Jun in these cell lines and found that crizotinib decreased the levels of both p-c-Jun and c-Jun in both HCC827/ER and HCC827/AR cells, but not in HCC827 cells (Fig. 5C). This result is consistent with its effect on suppressing AP-1-dependent DR4 transcription.
Fig. 5.
Crizotinib decreases DR4 mRNA levels (A), suppresses AP-1-dependent DR4 promoter activity (B) and decreased the levels of c-Jun and p-c-Jun (C) in EGFR-TKI resistant HCC827 cells. A, The indicated cell lines were treated with 0.5 μM crizotinib for 8 h and then harvested for preparation of total cellular RNA and subsequent RT-PCR. B, HCC827/AR cells were transfected with the given DR4 reporter plasmids for 18 h and then treated with DMSO or 0.5 μM crizotinib for additional 10 h. The cells were then harvested for luciferase activity assay. The data are means ± SDs of triplicate determinations. C, The indicated cancer cell lines were treated with different concentrations of MET inhibitors as indicated for 8 h and then harvested for preparation of whole-cell protein lysates and subsequent Western blot analysis. LE, longer exposure. D, The pathways by which a MET inhibitor (METi) or its combination with an EGFR inhibitor (EGFRi) suppresses DR4 expression in EGFR-TKI resistant HCC827 cells were illustrated.
The combination of osimertinib and crizotinib enhances suppression of ERK/c-Jun signaling and reduction of DR4 expression accompanied with augmented induction of apoptosis in MET-amplified osimertinib-resistant NSCLC cells
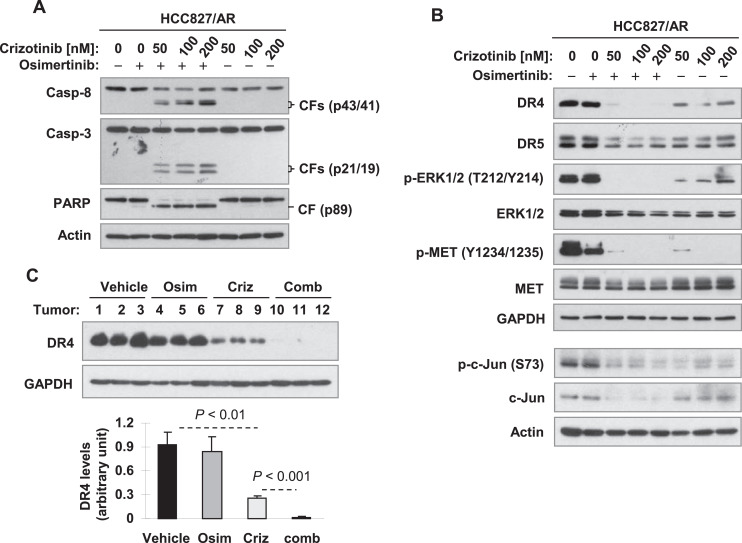
We previously showed that co-inhibition of MET overcomes acquired resistance of MET-amplified EGFR-TKI resistant NSCLC cells and tumors to osimertinib [4]. We then analyzed how the combination of osimertinib with crizotinib modulates DR4 expression in HCC827/AR cells under the condition that the combination enhances apoptosis. In this study, we could reproduce our previous result by detecting augmented cleavage of PARP and caspase-3, hallmarks of apoptosis, in HCC827/AR cells treated by the combination of osimertinib and crizotinib in comparison with cells exposed to each agent alone (Fig. 6A). Crizotinib alone suppressed ERK phosphorylation accompanied with decreased expression of both DR4 and DR5 in HCC827/AR cells; however, it enhanced the suppression of ERK phosphorylation and the reduction of DR4, but not DR5 expression, when combined with osimertinib (Fig. 6B). In addition, the combination also enhanced reduction of c-Jun levels in compared with each agent alone (Fig. 6B). Furthermore, we generated similar results with HCC827/AR xenografts, which showed that the combination of osimertinib and crizotinib was significantly more potent than both osimertinib and crizotinib alone in decreasing DR4 levels, although crizotinib alone also significantly reduced DR4 levels (Fig. 6C). Therefore, it is apparent that the combination augments induction of apoptosis and inhibition of the growth of osimertinib-resistant HCC827/AR cells and tumors accompanied with augmented suppression of DR4 expression.
Fig. 6.
The combination of osimertinib and crizotinib augments induction of apoptosis (A) accompanied with enhanced suppression of ERK/c-Jun signaling and reduction of DR4 (B) in HCC827/AR cells and enhanced DR4 reduction in HCC827/AR xenografts (C). A and B, HCC827/AR cells were exposed to 100 nM osimertinib (Osim), different concentrations of crizotinib (Criz) as indicated and their respective combinations for 10 h (B) or 24 h (A) and then harvested for preparation of whole-cell protein lysates and subsequent Western blotting for detection of the indicated proteins. CF, cleaved from. C, The indicated proteins in different tissue lysates were detected with Western blotting. The tumor samples were from the experiment reported previously [4].
Crizotinib and TRAIL combination enhances killing of MET-amplified HCC827/AR cells
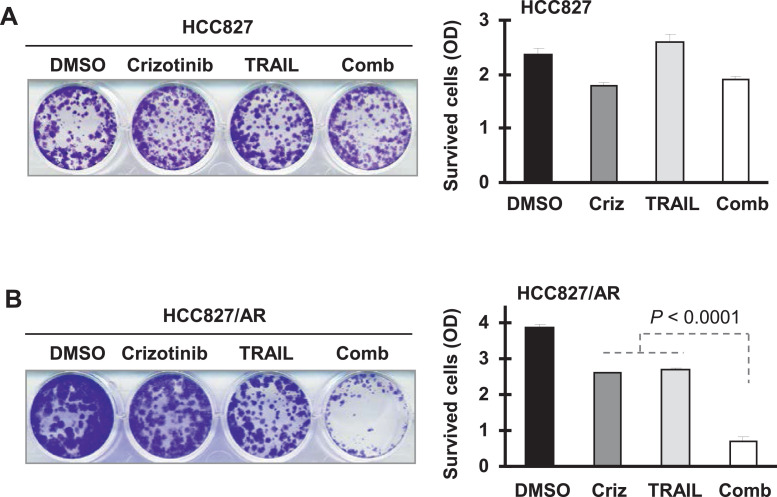
Since DR4 is the receptor for TRAIL, we further examined the effects of crizotinib and TRAIL combination on the growth of HCC827/AR cells in comparison with HCC827 cells. Considering that HCC827 cells are insensitive to TRAIL, we conducted a colony formation assay that allows us to repeatedly treat the cells with the tested agents for their long-term activities. HCC827 cells were insensitive to TRAIL and even the combination of crizotinib and TRAIL (Fig. 7A). However, HCC827/AR cells were sensitive to the combination although they responded in part to either agent (Fig. 7B). Hence, the combination of crizotinib and TRAIL effectively kills MET-amplified HCC827/AR cells.
Fig. 7.
The combination of crizotinib and TRAIL enhances suppression of colony growth of HCC827/AR cells (B), but not HCC827 cells (A). The indicated cell lines were plated in 12-well plates and treated on the second day with DMSO, 50 nM crizotinib (Criz), 20 ng/ml TRAIL or the combination of crizotinib and TRAIL. The treatments were repeated every 3 d. After 12 d, the colony was stained with crystal violet and photographed. Each column represents a mean ± SD of triplicate determinations.
Discussion
Both HCC827/ER and HCC827/AR cells have hyper-activated MET evidenced by very high levels of phosphorylated MET protein as demonstrated in our previous [4,22] and current study. Therefore, it makes sense that these cell lines were more sensitive to MET inhibitors than their parental cells and other NSCLC cells with no MET gene amplification. It is known that MET activates the MEK/ERK signaling pathway. In this study, both crizotinib and SGX523 effectively suppressed ERK phosphorylation accompanied with suppression of DR4 expression in HCC827/ER and HCC827/AR cells, but not in HCC827 cells, indicating the tight association between MEK/ERK suppression and DR4 downregulation. Moreover, crizotinib decreased DR4 mRNA levels, suppressed AP-1-dependent DR4 gene transcription and decreased the levels of both c-Jun and phosphorylated c-Jun. Hence, it is very likely that MET/MEK/ERK positively regulates DR4 expression through c-Jun/AP-1-dependent transcriptional regulation; upon MET inhibition, DR4 expression is suppressed (Fig. 5D). Our findings in this study provide additional evidence supporting our previous report that the MEK/ERK/AP-1 signaling positively regulates DR4 expression [16].
In HCC827 cells, both crizotininb and SGX523 suppressed basal levels of p-MET, although its levels were relatively low; however, they were unable to inhibit ERK phosphorylation. This may suggest that the basal levels of p-ERK is controlled by other signaling pathways independent of MET in this cell line. Hence, the poor response of HCC827 cells to MET inhibitors can be explained by the dispensable role of MET in controlling MEK/ERK signaling in this cell line. Once they become resistant due to MET amplification and protein hyperactivation, then MET is indispensable in activating the MEK/ERK signaling. Accordingly, these cell lines (e.g., HCC827/ER and HCC827/AR) respond well to MET inhibitors as demonstrated in this study and to the combination of MET inhibition and osimertinib as demonstrated previously [4,22].
We noted that crizotinib increased DR4 levels in HCC827 and other NSCLC cell lines without mutant EGFR. Since crizotinib did not or minimally suppressed ERK phosphorylation, it is likely that crizotinib increases DR4 in these cell lines through different and yet-to-be identified mechanisms. Beyond DR4, it also upregulated DR5 in these cell lines. Nonetheless, these findings warrant future study to demonstrate the mechanisms by which crizotinib increases the expression of DR4 and DR5 in these cell lines and its impact on TRAIL-induced apoptosis.
DR5 expression is also positively regulated by the Raf/MEK/ERK signaling pathway as we demonstrated previously [14,15]. However, the underlying mechanism involves CHOP/ILK-mediated gene transcription. Although JNK/c-Jun activation enhances the transcription, MET inhibitors did not or minimally suppressed DR5 expression in MET-amplified EGFR-TKI resistant HCC827 cell lines. These findings demonstrate the distinct mechanisms underlying MEK/ERK-dependent regulation of DR4 and DR5 transcription.
In this study, ARQ197 behaved differently from both crizotinib and SGX523 in suppressing MET/ERK signaling and modulating DR4 expression in both HCC827 and HCC827/AR cells as it did not suppress the phosphorylation of MET and ERK and rather enhanced DR4 expression. In this study, we have validated the findings on MET inhibition by small molecule MET inhibitors (e.g., crizotinib and SGX523) and suppression of DR4 expression by genetic knockdown of MET expression in both HCC827/ER and HCC827/AR cells. Therefore, we suggest that ARQ197 may not be an ideal MET inhibitor.
It was reported that silencing of DR5, but not DR4, in Huh-7 hepatocellular carcinoma cells attenuates TRAIL-induced apoptosis [23]. We previously reported that knockdown of DR4 enhanced apoptosis induced by TRAIL or the combination of TRAIL and GGTI-298 (a geranylgeranyltransferase I inhibitor) in NSCLC cells, whereas DR5 silencing abolished apoptosis induced by TRAIL or the GGTI-298 and TRAIL combination [13]. In this study, DR4 downregulation by MET inhibitors occurred in the sensitive MET-amplified NSCLC cells. The combination of crizotinib and osimertinib enhanced suppression of DR4 expression accompanied with augmented induction of apoptosis in HCC827/AR cells. This combination also enhanced DR4 reduction in HCC827/AR xenograft tumors while effectively inhibiting the growth of these tumors in vivo [4]. Consistently, the combination of crizotinib and TRAIL enhanced the killing of HCC827/AR cells, in which DR4 was decreased by crizotinib, but not of HCC827 cells, in which DR4 expression could not be suppressed by crizotinib. These results together suggest that DR4 may have an unrecognized anti-apoptotic function or a positive role in regulation of cancer cell growth under a given condition although we currently do not know the underlying mechanisms. However, our findings warrant further investigation in this direction.
Since DR4 reduction is tightly associated with cell response to MET inhibitors, the combination of a MET inhibitor with an EGFR-TKI or the combination of a MET inhibitor with TRAIL in MET-amplified EGFR-TKI resistant NSCLC cells, we suggest that DR4 reduction may serve as a predictive marker for these treatments. Moreover, any strategies that cause DR4 reduction may be effective in overcoming acquired resistance of EGFRm NSCLC cells or tumors to EGFR-TKIs due to MET amplification or in sensitizing certain cancer cells to TRAIL-induced apoptosis.
Authors' contributions
All authors contributed to the study conception and design. Experimental conduction, data collection and analysis were performed by Liang Deng, Karin A. Vallega, Shuo Zhang and Puyu Shi. The first draft of the manuscript was written by Liang Deng and Shi-Yong Sun and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Footnotes
Funding: This study was supported by NIH/NCIR01 CA223220 (to SYS), R01 CA245386 (to SYS), UG1 CA233259 (to SYS) and Lung Cancer SPOREP50 CA217691 DRP award (to S-YS) and Emory University Winship Cancer Institute lung cancer pilot funds (to SYS).
Conflicts of interest: All authors declare no conflicts of interest.
References
- 1.Tartarone A, Lerose R. Clinical approaches to treat patients with non-small cell lung cancer and epidermal growth factor receptor tyrosine kinase inhibitor acquired resistance. Therap Adv Respir Dis. 2015;9:242–250. doi: 10.1177/1753465815587820. [DOI] [PubMed] [Google Scholar]
- 2.Juchum M, Gunther M, Laufer SA. Fighting cancer drug resistance: opportunities and challenges for mutation-specific EGFR inhibitors. Drug Resist Updat Rev Comment Antimicrob Anticancer Chemother. 2015;20:10–28. doi: 10.1016/j.drup.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Remon J, Moran T, Majem M, Reguart N, Dalmau E, Marquez-Medina D, Lianes P. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer: a new era begins. Cancer Treat Rev. 2014;40:93–101. doi: 10.1016/j.ctrv.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Shi P, Oh YT, Zhang G, Yao W, Yue P, Li Y, Kanteti R, Riehm J, Salgia R, Owonikoko TK. Met gene amplification and protein hyperactivation is a mechanism of resistance to both first and third generation EGFR inhibitors in lung cancer treatment. Cancer Lett. 2016;380:494–504. doi: 10.1016/j.canlet.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, Kurtz DM, Stehr H, Scherer F, Karlovich CA. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nature communications. 2016;7:11815. doi: 10.1038/ncomms11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ou SH, Agarwal N, SM Ali. High MET amplification level as a resistance mechanism to osimertinib (AZD9291) in a patient that symptomatically responded to crizotinib treatment post-osimertinib progression. Lung Cancer. 2016;98:59–61. doi: 10.1016/j.lungcan.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Ortiz-Cuaran S, Scheffler M, Plenker D, Dahmen I, Scheel A, Fernandez-Cuesta L, Meder L, Lovly CM, Persigehl T, Merkelbach-Bruse S. Heterogeneous mechanisms of primary and acquired resistance to third-generation EGFR inhibitors. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-15-1915. [DOI] [PubMed] [Google Scholar]
- 8.Yang A, Wilson NS, Ashkenazi A. Proapoptotic DR4 and DR5 signaling in cancer cells: toward clinical translation. Curr Opin Cell Biol. 2010;22:837–844. doi: 10.1016/j.ceb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 9.van Roosmalen IA, Quax WJ, Kruyt FA. Two death-inducing human TRAIL receptors to target in cancer: similar or distinct regulation and function? Biochem Pharmacol. 2014;91:447–456. doi: 10.1016/j.bcp.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Falschlehner C, Schaefer U, Walczak H. Following TRAIL's path in the immune system. Immunology. 2009;127:145–154. doi: 10.1111/j.1365-2567.2009.03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer. 2008;8:782–798. doi: 10.1038/nrc2465. [DOI] [PubMed] [Google Scholar]
- 12.OR E, Tirincsi A, Logue SE, Szegezdi E. The Janus face of death receptor signaling during tumor immunoediting. Frontiers in immunology. 2016;7:446. doi: 10.3389/fimmu.2016.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S, Fu L, Raja SM, Yue P, Khuri FR, Sun SY. Dissecting the roles of DR4, DR5 and c-FLIP in the regulation of geranylgeranyltransferase I inhibition-mediated augmentation of TRAIL-induced apoptosis. Mol Cancer. 2010;9:23. doi: 10.1186/1476-4598-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh YT, Liu X, Yue P, Kang S, Chen J, Taunton J, Khuri FR, Sun SY. ERK/ribosomal S6 kinase (RSK) signaling positively regulates death receptor 5 expression through co-activation of CHOP and Elk1. J Biol Chem. 2010;285:41310–41319. doi: 10.1074/jbc.M110.153775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh YT, Yue P, Zhou W, Balko JM, Black EP, Owonikoko TK, Khuri FR, Sun SY. Oncogenic Ras and B-Raf proteins positively regulate death receptor 5 expression through co-activation of ERK and JNK signaling. J Biol Chem. 2012;287:257–267. doi: 10.1074/jbc.M111.304006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao W, Oh YT, Deng J, Yue P, Deng L, Huang H, Zhou W, Sun SY. Expression of death receptor 4 is positively regulated by MEK/ERK/AP-1 signaling and suppressed upon MEK inhibition. J Biol Chem. 2016 doi: 10.1074/jbc.M116.738302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi P, Oh YT, Deng L, Zhang G, Qian G, Zhang S, Ren H, Wu G, Legendre B, Jr., Anderson E. Overcoming acquired resistance to AZD9291, A third-generation EGFR inhibitor, through modulation of MEK/ERK-dependent Bim and Mcl-1 degradation. Clin Cancer Res. 2017;23:6567–6579. doi: 10.1158/1078-0432.CCR-17-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun SY, Yue P, Dawson MI, Shroot B, Michel S, Lamph WW, Heyman RA, Teng M, Chandraratna RA, Shudo K. Differential effects of synthetic nuclear retinoid receptor-selective retinoids on the growth of human non-small cell lung carcinoma cells. Cancer Res. 1997;57:4931–4939. [PubMed] [Google Scholar]
- 19.Fu L, Lin YD, Elrod HA, Yue P, Oh Y, Li B, Tao H, Chen GZ, Shin DM, Khuri FR. c-Jun NH2-terminal kinase-dependent upregulation of DR5 mediates cooperative induction of apoptosis by perifosine and TRAIL. Mol Cancer. 2010;9:315. doi: 10.1186/1476-4598-9-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhandapani L, Yue P, Ramalingam SS, Khuri FR, Sun SY. Retinoic acid enhances TRAIL-induced apoptosis in cancer cells by upregulating TRAIL receptor 1 expression. Cancer Res. 2011;71:5245–5254. doi: 10.1158/0008-5472.CAN-10-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan B, Yue P, Lotan R, Sun SY. Evidence that the human death receptor 4 is regulated by activator protein 1. Oncogene. 2002;21:3121–3129. doi: 10.1038/sj.onc.1205430. [DOI] [PubMed] [Google Scholar]
- 22.Yu D, Li Y, Sun KD, Gu J, Chen Z, Owonikoko TK, Ramalingam SS, Sun SY. The novel MET inhibitor, HQP8361, possesses single agent activity and enhances therapeutic efficacy of AZD9291 (osimertinib) against AZD9291-resistant NSCLC cells with activated MET. Am J Cancer Res. 2020;10:3316–3327. [PMC free article] [PubMed] [Google Scholar]
- 23.Akazawa Y, Mott JL, Bronk SF, Werneburg NW, Kahraman A, Guicciardi ME, Meng XW, Kohno S, Shah VH, Kaufmann SH. Death receptor 5 internalization is required for lysosomal permeabilization by TRAIL in malignant liver cell lines. Gastroenterology. 2009;136:2365–2376. doi: 10.1053/j.gastro.2009.02.071. e2361-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]