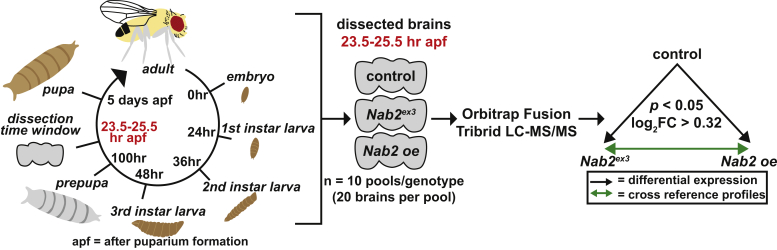
Figure 2.
Study design and analytic approach for quantitative proteomic analysis of Drosophila pupal brains. A workflow summary showing dissection window, experimental design, and analysis. The Drosophila life cycle with developmental stage and hours of development depicted with the dissection time window (23.25–25.5 h APF) in red, left. There were 600 developmentally timed brain samples that were pooled by the genotype, control (C155>Gal4, w1118); Nab2 zygotic null (Nab2ex3 = C155>Gal4;;Nab2ex3); and Nab2 overexpression in neurons (Nab2 oe = C155>Gal4;Nab2EP3716;Nab2ex3), and by sex, resulting in 30 individual pools, center. Each sample pool was processed, analyzed using an Orbitrap Fusion Tribrid Mass Spectrometer and quantified using MaxQuant against the Drosophila melanogaster UniProt database, center. Arrows depict the performed analyses. Differential protein abundance of Nab2ex3 and Nab2 oe brains was calculated with an FDR-adjusted p-value (black arrows) and then second-degree analyses cross-referencing the Nab2ex3 and Nab2 oe proteomic profiles (green arrows), right. APF, after puparium formation; FDR, false discovery rate.