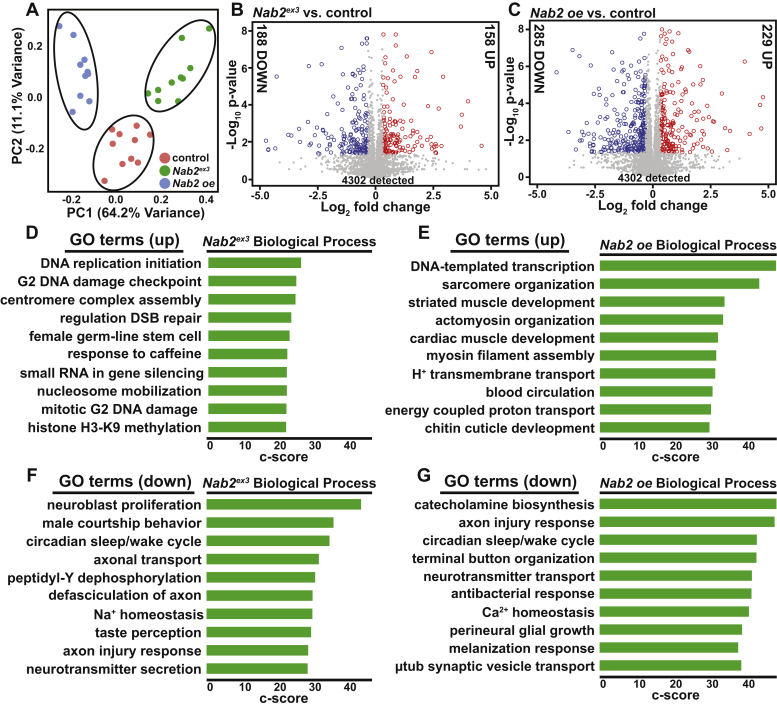
Figure 3.
Quantitative proteomic analysis of developmentally timed pupal brains reveals a role for Nab2 in neurodevelopment.A, principal component analysis (PCA) of proteomic data from 24 h after puparium formation Drosophila brains from ten biological replicates of control, Nab2ex3, and Nab2 oe flies (control = C155>Gal4, w1118; Nab2ex3 = C155>Gal4;;Nab2ex3; Nab2 oe =C155>Gal4;Nab2EP3716;Nab2ex3) show results cluster based on the genotype and that Nab2ex3 and Nab2 oe are distinct from control and each other. PCA was performed in RStudio using prcomp (default stats package v3.5.1), and summed peptide intensities were used as the input. B and C, volcano plots show proteins differentially expressed in each Nab2 genotype compared with the control [B, Nab2ex3 (346; 188 down and 158 up) and C, Nab2 oe compared with the control (514; 285 down and 229 up)]. Ten biological replicates (n = 10) per genotype (20 brains per pooled biological replicate) with 30 technical replicates in total. Significance thresholds: log2(≥0.32 and ≤−0.32) and −log10(p-value) ≥1.3; thresholds were based on power calculation and instrumental limits. Protein abundance change (down or up) indicated on each side of the plot (log2Nab2ex3/Cont or Nab2 oe/Cont: gray = not significant, blue ≤ −0.32, red ≥ 0.32). The number of differentially expressed proteins to total detected proteins is shown atop the graph; 346 of 4302 Nab2ex3 proteins are differentially expressed (B) and 514 of 4302 Nab2 oe proteins are differentially expressed (C). D–G, the enriched gene ontology terms from FlyEnrichr database for biological process are shown for proteins increased log2(Nab2ex3/Cont) ≥0.32 in panel D Nab2ex3 and log2(Nab2 oe/Cont) in panel E Nab2 oe and decreased log2(Nab2ex3/Cont) ≤−0.32 in panel F Nab2ex3 and log2(Nab2 oe/Cont) in panel G Nab2 oe. The bars shown correspond to the top ten c-scores (c-score = ln(adj p-val) ∗ z-score) in each dataset (adjusted p-value <0.05) (74, 75, 76).