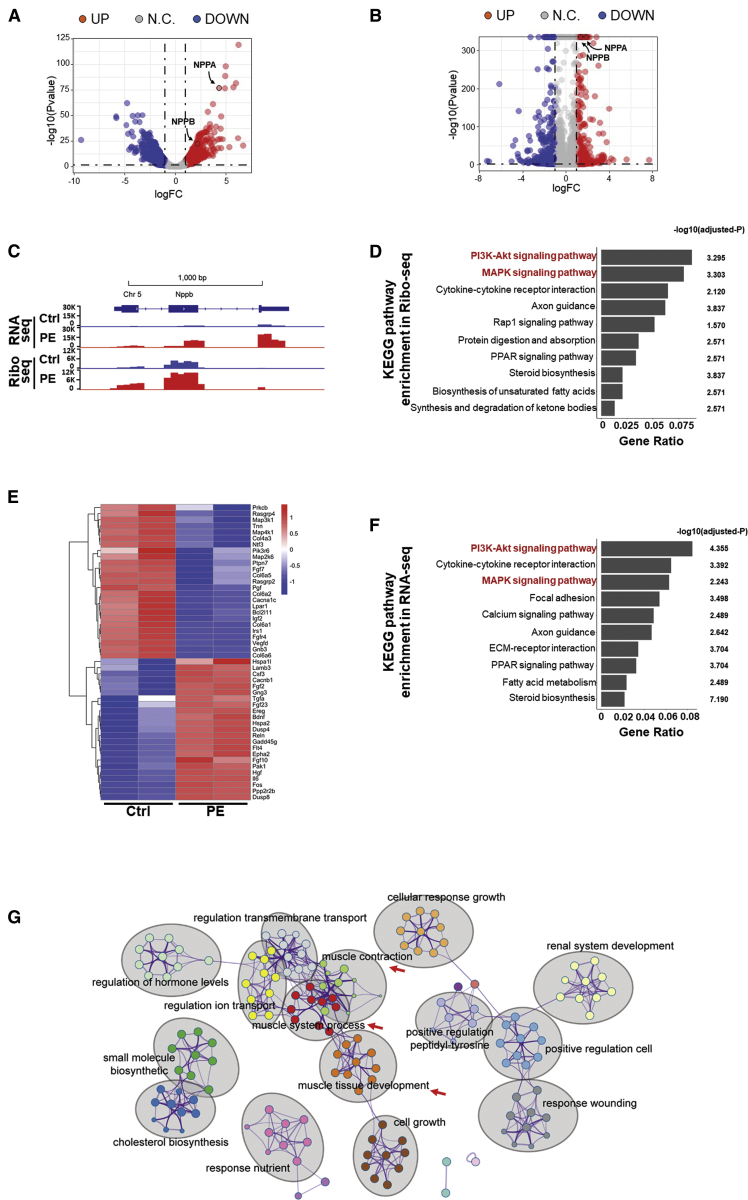
Figure 2.
Integrative genome-wide analyses of Ribo-seq and RNA-seq from PE-treated cardiomyocytes
(A and B) Volcano maps of dys-regulated genes in RNA-seq (A) and Ribo-seq (B). Red and blue dots represent the upregulated and downregulated genes, respectively (FDR < 0.05; |Log2(fold change)| > 1). (C) The genomic view of reads in RNA-seq and Ribo-seq for hypertrophic marker gene Nppb in the IGV browser. y axis shows the mapped reads number. (D) KEGG pathway analysis of significantly dys-regulated genes in translation (Ribo-seq). Enriched pathways are ranked by gene ratio. (E) Heatmap of dys-regulated genes in the “PI3K/AKT signaling pathway” and “MAPK signaling pathway” from Ribo-seq. (F) KEGG pathway analysis of significantly dys-regulated genes in transcription (RNA-seq). Enriched pathways are ranked by gene ratio. (G) Metascape analysis of PE-treated cardiomyocyte transcriptome. Enriched ontology clusters were colored by cluster ID and shown by gray ovals. Clusters described in the main text are indicated by red arrows.