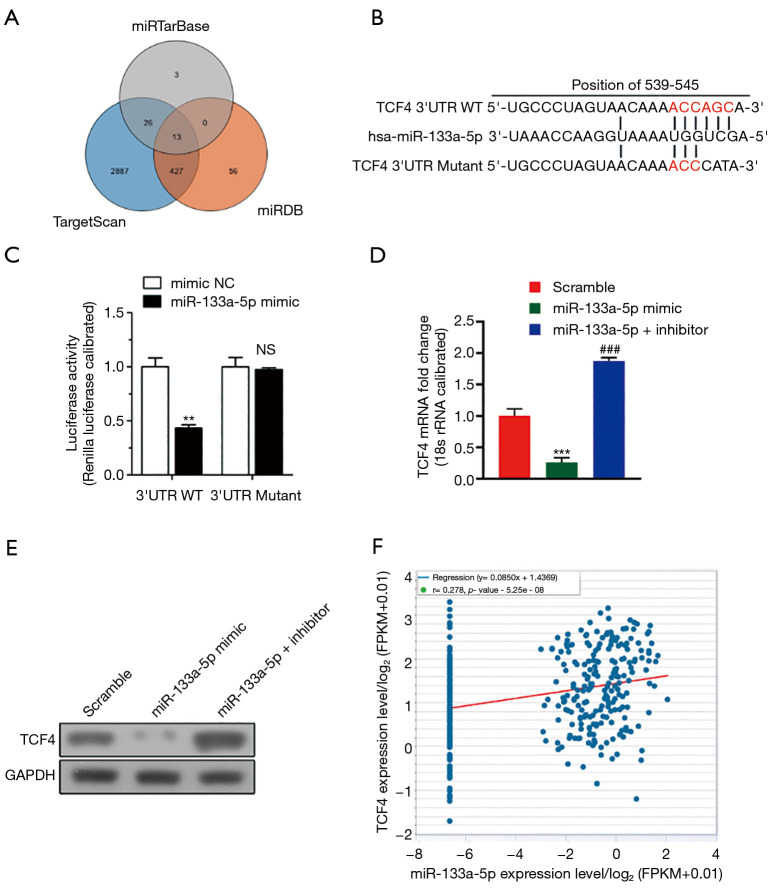
Figure 3.
Screen the target genes of MiR-133a-5p by RT-QPCR by bioinformatic analysis and in vitro assay. (A) Veen plot showed the common targets of the MiR-133a-5p was screened by the three databases: miRTarBase, miRDB, Target Scan. (B) The binding sites of TCF4 and MiR-133a-5p. (C) Effects of miR-133a-5p mimics the activity of wild-type, and mutant 3’-UTRs of TCF4 were analyzed by dual-luciferase reporter assay. (D) The relative TCF4 mRNA levels of predicted target genes in cells transfected with miR-133a-5p mimics and negative control miRNA. (E) The relative TCF4 protein levels of predicted target genes in cells transfected with miR-133a-5p inhibitor and negative control miRNA. GAPDH functioned as the loading control. (F) The scattering plot shows the correlation expression between the TCF4 expression levels and miR-133a-5p. One-way ANOVA analysis. NS, not significant. **P<0.01 and ***P<0.001. ###P<0.001.