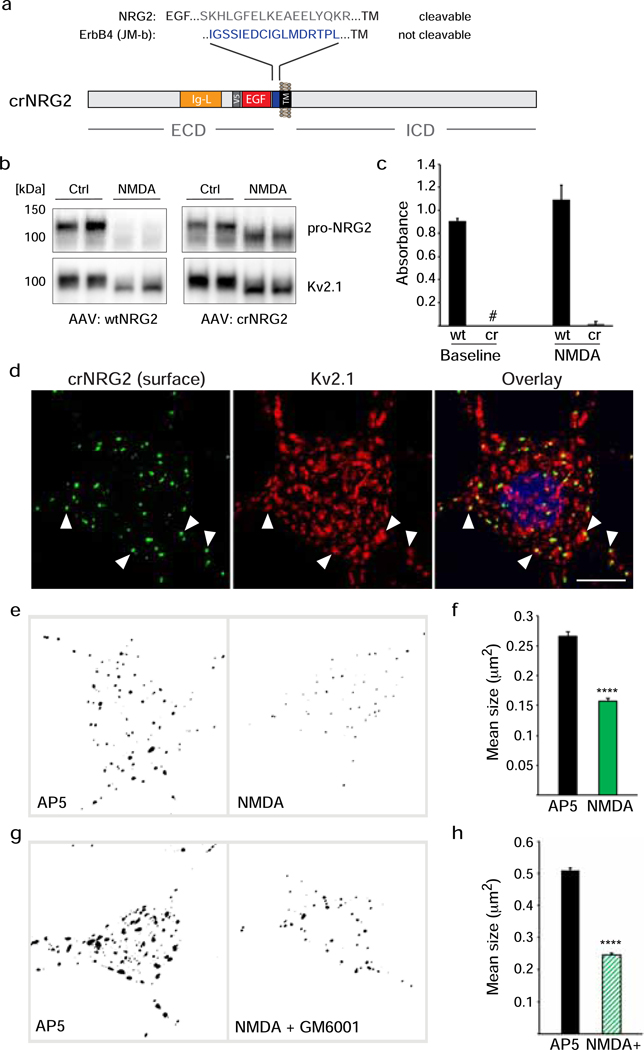
Fig. 2. Cleavage-resistant NRG2 reveals NMDAR-dependent downregulation of pro-NRG2 at ER-PM junctions in the absence of ectodomain shedding.
(a) Diagram of cleavage-resistant NRG2 (crNRG2), showing the replaced juxtamembrane extracellular sequence of wtNRG2 and the inserted non-cleavable juxtamembrane sequence of ErbB4 JM-b (see also [36]). Ig-like (Ig-L) and EGF-like domains are also indicated, as well as the location of a V5 epitope tag present in all NRG constructs described in this study that was used for ELISA and immunofluorescence cytochemistry. (b) Western blots of lysates prepared from hippocampal neurons expressing wtNRG2 (left) or crNRG2 (right), showing the effects of 10 min of 50 μM NMDA on pro-NRG2 protein levels. Note that NMDAR stimulation has little, if any, effect on crNRG2 protein levels (top) but causes a downward shift in its apparent electrophoretic mobility that parallels the shift in Kv2.1 mobility (below). (c) ELISA of ecto-NRG2 in the corresponding culture supernatants, demonstrating ectodomain shedding of wtNRG2 but not crNRG2. The (#) denotes that levels were below the limits of detection. Data represent the means of two biological replicates. (d) Representative confocal Z-projection of a hippocampal neuron expressing crNRG2 (surface-labeled with anti-V5), showing colocalization with endogenous Kv2.1 clusters. DAPI was included in the overlay image on the right. Arrowheads point to examples of closely associated signals for crNRG2 and Kv2.1. Scale bar = 10 μm (e) Representative binary images of crNRG2 puncta in hippocampal neurons following 10-min treatments with 100 μM AP5 (left) or 50 μM NMDA (right). (f) Quantitative analysis reveals reduction of mean crNRG2 puncta size after NMDAR stimulation (n=752 puncta from 11 neurons (AP5) and 397 puncta from 12 neurons (NMDA)). (g,h) Similar analysis of neurons expressing wtNRG2 and treated with AP5 or NMDA plus GM6001 (10 μM) (n=846 puncta from 12 neurons (AP5) and 566 puncta from 12 neurons (NMDA+GM6001)). ****, p<0.0001 (unpaired t-test).