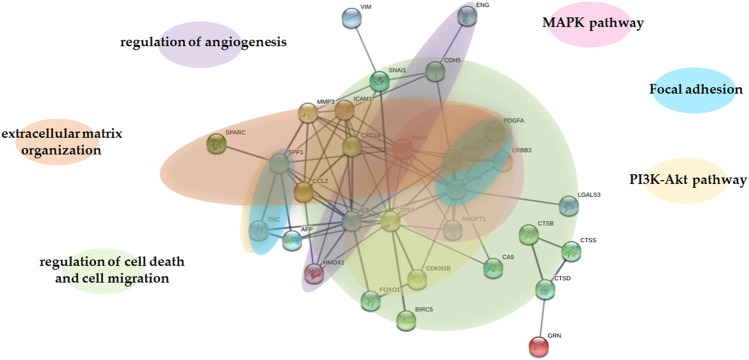
FIGURE 5.
STRING analysis of proteins in WM278 and WM1617 cells. STRING database version 11.0 (http://string-db.org) was used to determine the protein-protein interactions of the 38 proteins differentially expressed in the original and transfected cells. Interactions predicted with high confidence level: 0.700 were included in the analyses, and proteins with no predicted interactions were removed. STRING: Search Tool for the Retrieval of Interacting Genes; AFP: alpha-fetoprotein; ANGPT1: angiopoietin-1; BIRC5: survivin; CA9: carbonic anhydrase IX; CCL2: C-C motif chemokine two; CDH5: VE-cadherin; CDKN1B: cyclin-dependent kinase inhibitor 1B/p27; CTSB: cathepsin B; CTSD: cathepsin D; CTSS: cathepsin S; CXCL8: interleukin-8; EGFR: epidermal growth factor receptor; ENG: endoglin; ERBB2: receptor tyrosine-protein kinase erbB-2; ERBB3: receptor tyrosine-protein kinase erbB-3; FGF2: basic fibroblast growth factor; FOX01: forkhead box protein O1; GRN: progranulin; HMOX1: heme oxygenase one; ICAM1: intercellular adhesion molecule one; IL-6: interleukin-6; LGALS3: galectin-3; MMP3: matrix metallo-proteinase-3; PDGFA: platelet-derived growth factor AA; SNAI1: snail; SPARC: secreted protein acidic and rich in cysteine; SPP1: osteopontin; TNC: tenascin C; TP53: cellular tumor antigen p53; VIM: vimentin; MAPK pathway: The Mitogen-Activated Protein Kinase Pathway; PI3K-Akt pathway: phosphatidylinositol 3-kinase protein kinase B signaling pathway.