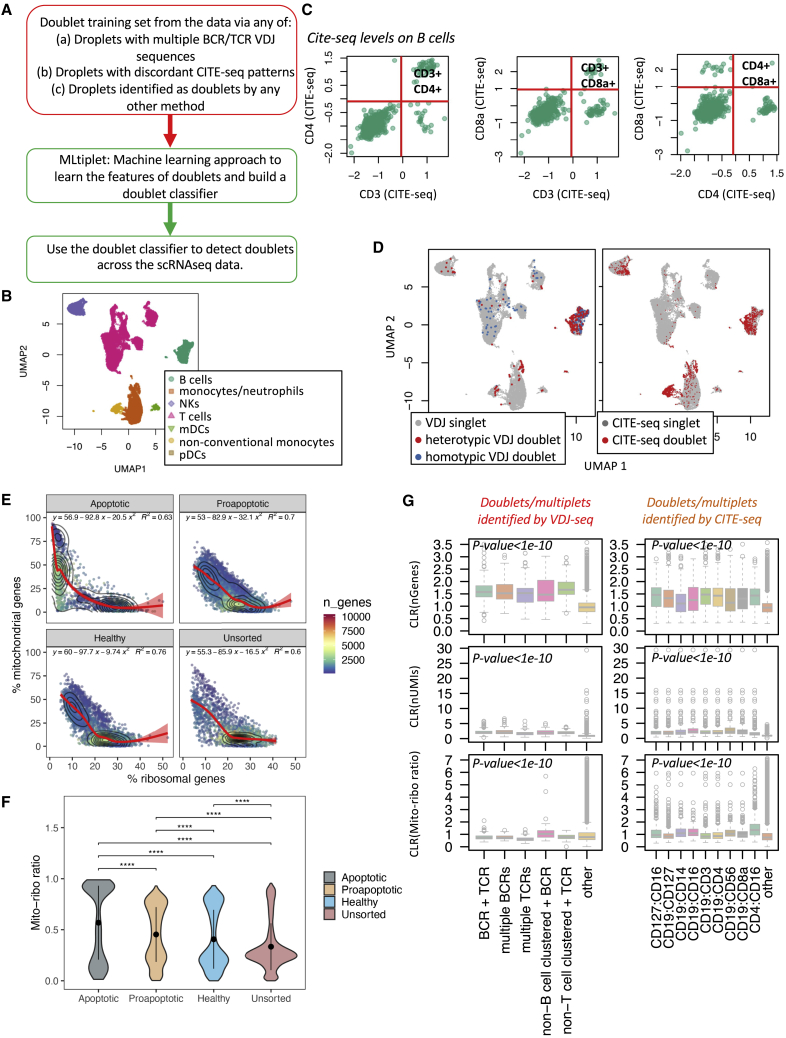
Figure 1.
Multi-omics aids the identification of doublets and/or multiplets
(A) Schematic of approach to identify scRNA-seq doublets and/or multiplets using the CITE-seq and VDJ modalities. Droplets with a transcriptome resembling non-B or non-T cells that captured BCR or TCR sequences, respectively, were considered as potential doublets and/or multiplets.
(B) Uniform manifold approximation and projection (UMAP) dimensionality reduction of three healthy PBMC datasets colored by cell type.
(C) Examples of the CITE-seq levels between CD3, CD4, and CD8 for the three individuals for the B cell cluster, with the red lines corresponding to the CITE-seq positivity thresholds.
(D) UMAP dimensionality reduction of three healthy PBMCs colored by VDJ doublets (left, co-capture of discordant VDJs) or CITE-seq doublets (right, co-capture of the corresponding mutually exclusive CITE-seq pair). Homotypic doublets were defined as those containing multiple BCRs or multiple TCRs, and the heterotypic doublets are defined as the remainder (droplets containing both BCR(s) and TCR(s) or droplets containing BCRs or TCRs that do not have transcriptional profiles that resemble B or T cells, respectively).
(E) Generalized additive models fitted on percentage mitochondrial genes versus percentage ribosomal genes, R2 values shown top right; from the HEK293 dataset.
(F) Mito-ribo ratio values per enriched HEK293 populations. p values were calculated across groups by using Wilcoxon test with Bonferroni correction for pairwise comparisons.
(G) The relative numbers of genes (nGenes), number of RNA molecules (nUMI), mito-ribo ratio (mitoribo_ratio), and nUMIs_VDJ per droplet for the VDJ-identified doublets and/or multiplets (left) and the CITE-seq identified doublets and/or multiplets (right). “Other” refers to droplets that were not identified as doublets and/or multiplets from the VDJ-seq or CITE-seq data. The p values of the differences between the feature distributions of the doublet/multiplets detected and the remainder of the droplets provided (two-sided Wilcoxon test). ∗∗∗∗p < 0.00005 (Wilcoxon test). Abbreviation is as follows: CLR, centered log-ratio transformed.