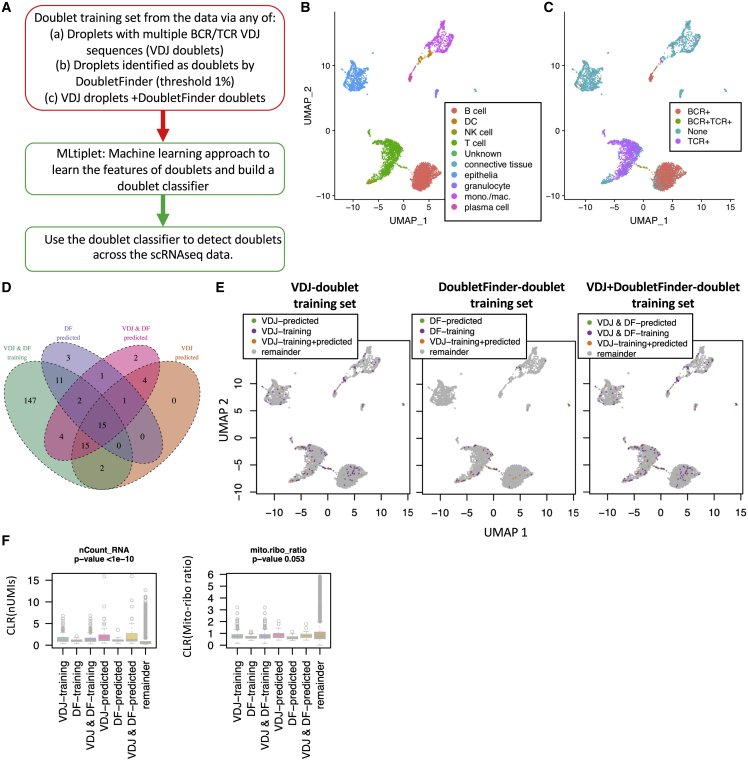
Figure 4.
Validation of MLtiplet on an NSCLC tumor dataset
Doublet detection on a non-small cell lung cancer (NSCLC) dataset.
(A–C) Shown are the (A) schematic of the training datasets for doublet/multiplet prediction by MLtiplet. UMAP plots of (B) the annotated cell types and (C) VDJ-seq heterotypic doublets.
(D) Venn diagram showing the numbers of droplets used as the combined identified doublets and/or multiples using both DoubletFinder and VDJ-seq (green), and the predicted doublets and/or multiplets from MLtiplet using the DoubletFinder-derived training dataset (blue), VDJ-seq-derived training dataset (orange), and DoubletFinder plus VDJ-seq-derived training dataset (pink).
(E) UMAP plots of the training and predicted doublets and/or multiples using each approach.
(F) The relative numbers of RNA molecules (nUMI) and mito-ribo ratio (mitoribo_ratio) per cell for the VDJ-identified doublets and/or multiplets, CITE-seq-identified doublets and/or multiplets, MLtiplet-predicted doublets and/or multiplets, and the remainder (predicted singlets by MLtiplet).