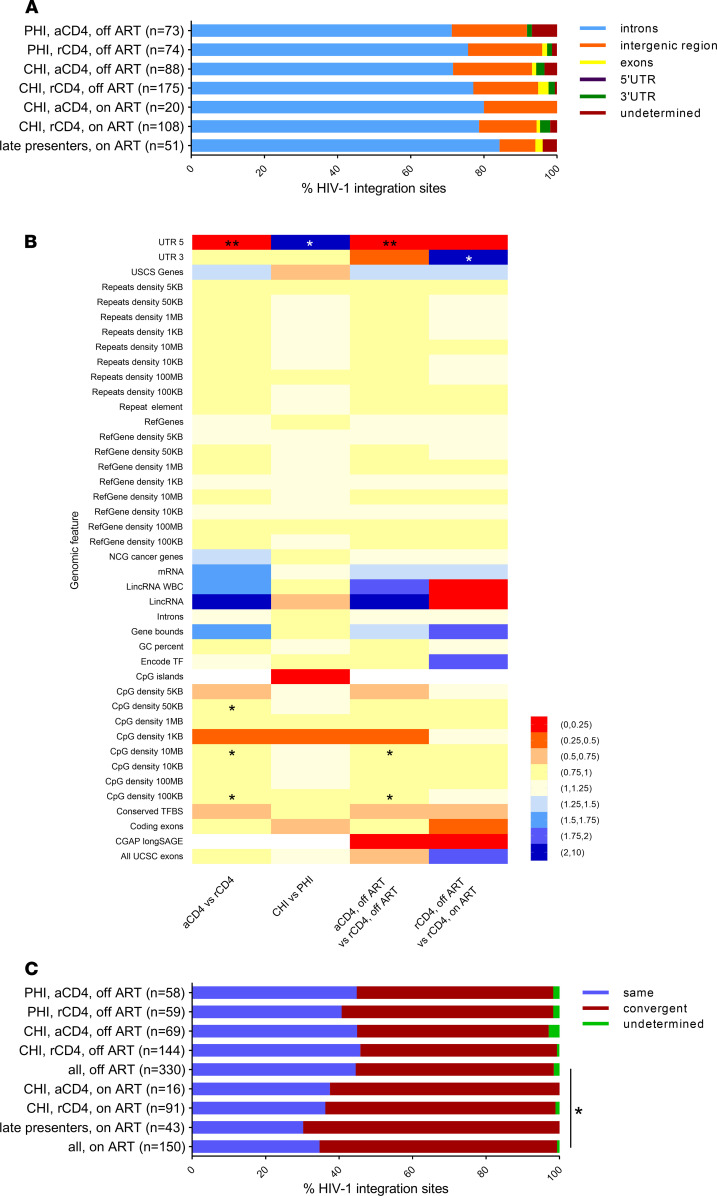
Figure 1. Genomic features of HIV-1 integration site libraries from longitudinally sampled resting and activated CD4+ T cells obtained from HIV-1–infected individuals during primary and chronic HIV-1 infection and during periods on and off ART and PBMCs from late presenters.
(A) Genomic distribution of HIV-1 integration sites. Undetermined HIV-1 integration sites were caused by splice variants. (B) Pairwise comparison of genomic features associated with HIV-1 integration sites in the different cell populations. The Benjamini-Hochberg method was used to correct for multiple hypothesis testing. (C) The orientation of HIV-1 proviruses relative to the host gene transcription orientation. Only intragenic HIV-1 integration sites were considered. P values were calculated using the 2-tailed Fisher’s exact test: *, P < 0.05, **, P < 0.01.