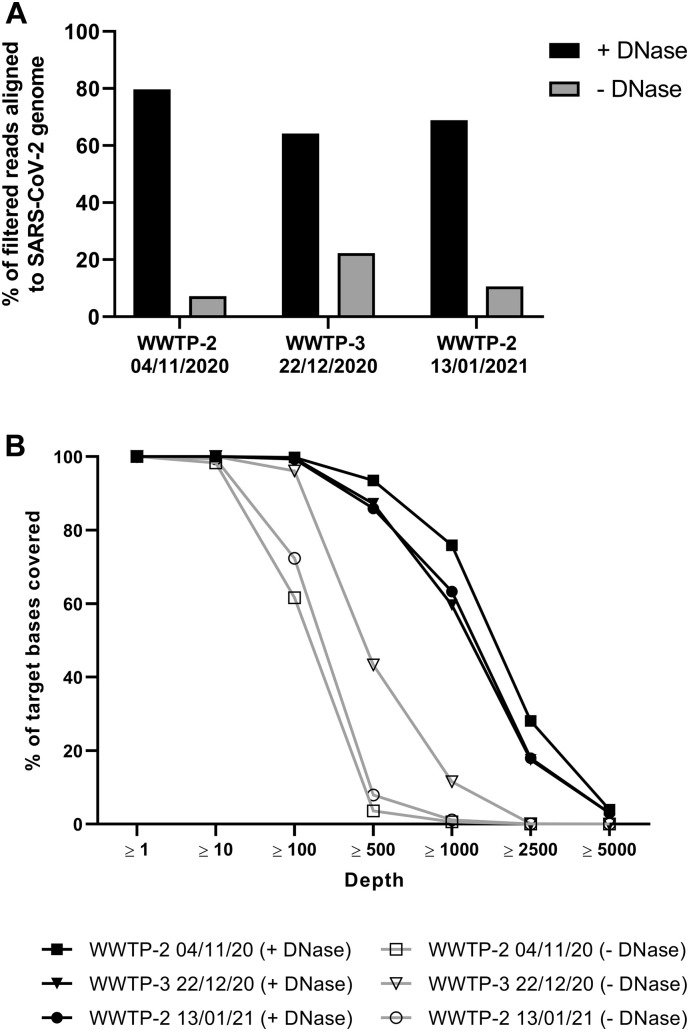
Fig. 8.
Analysis of wastewater nucleic acid for SARS-CoV-2 sequencing. Nucleic acid from the three wastewater collection sites and a pooled sample from the three collection sites at the indicated timepoints was used to generate SARS-CoV-2 amplicon libraries. Each dataset was randomly subsampled to 650,000 reads for comparison. (A) Percent of filtered reads aligned to SARS-CoV-2 genome for DNase-treated samples (+ DNase) or untreated total nucleic acid samples (− DNase). (B) Percent of target bases on the SARS-CoV-2 genome covered at or above indicated depths.