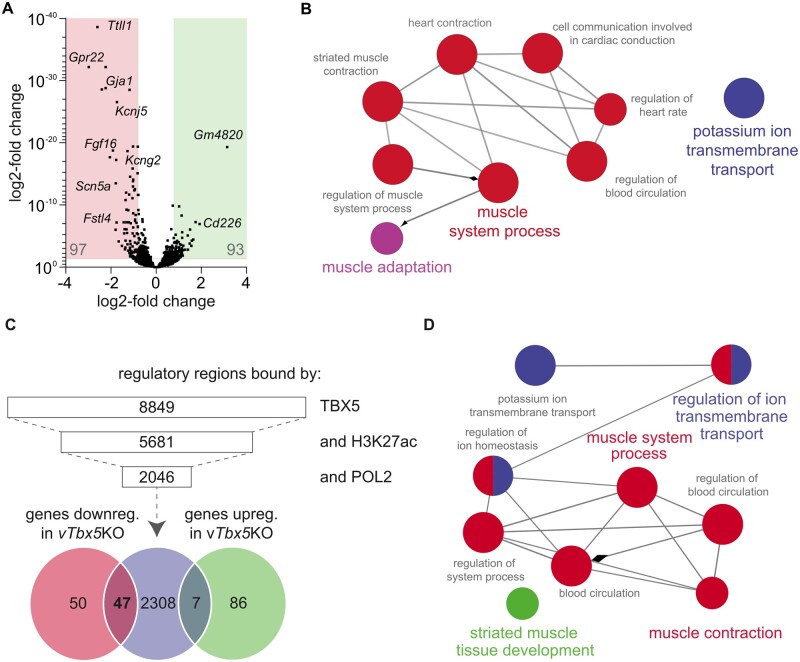
Figure 5.
Integrative chromatin occupancy and transcriptome analysis identified novel putative TBX5 downstream targets in the adult ventricle. (A) RNA-Sequencing results displayed in a volcano plot. Ninety-seven genes are down- and 93 are up-regulated in vTbx5KO ventricles using a cut-off of P < 0.05 and log2-fold-change of >0.8 or <−0.8. (B) Gene ontology analysis (ClueGO32,33) of biological processes of the down-regulated genes in vTbx5KO mice. (C) TBX5 co-occupancy with marks of active enhancers H3K27ac and POL2 identified 2046 putative TBX5 active enhancers. These were annotated to genes with GREAT (Gja1 and Fgf16 enhancers were not annotated automatically by GREAT, but manually identified by BED file analysis in the IGV platform34,35). Venn-diagram shows the intersection of those genes with the regulated transcripts of the RNA-Seq analysis. (D) Gene ontology analysis of biological processes for 47 ventricular TBX5 targets.