Extended Data Figure 3: Increased sensitivity of aneuploid cancer cells to SACi remains significant when associated genomic and phenotypic features are controlled for.
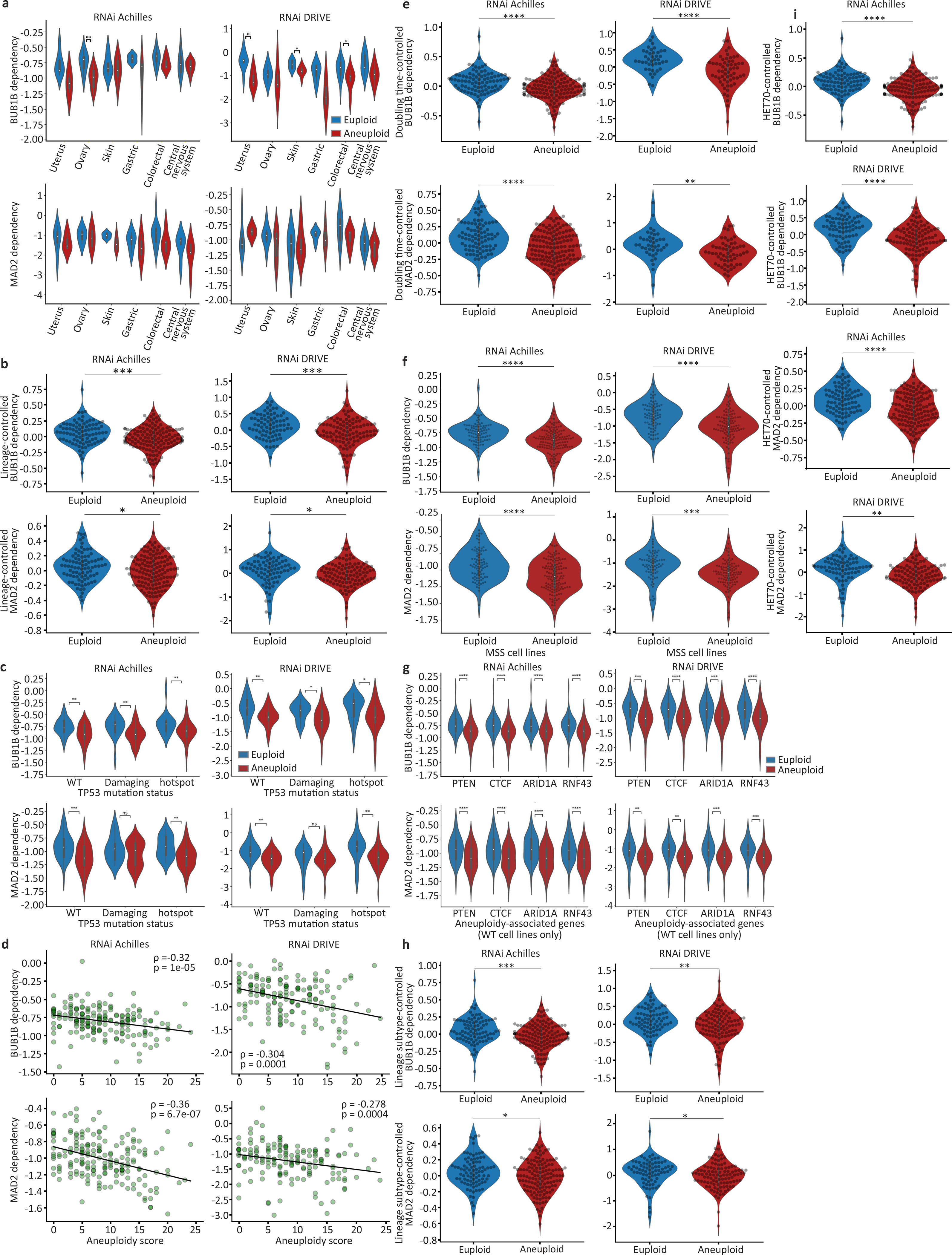
(a) The sensitivity of near-euploid and highly-aneuploid cancer cell lines to the knockdown of BUB1B (top) and MAD2 (bottom) in the Achilles (left) and DRIVE (right) RNAi screens across multiple cell lineages. *, p<0.05; **, p<0.01; two-tailed t-test. (b) The sensitivity of near-euploid and highly-aneuploid cancer cell lines to the knockdown of BUB1B (top) and MAD2 (bottom) in the Achilles (left) and DRIVE (right) RNAi screens, after accounting for lineage-specific differences in gene dependency scores using linear regression. ***, p=2e-04; * p=0.013; for RNAi-Achilles BUB1B and MAD2 dependencies, respectively; ***, p=5e-04; *, p=0.044; RNAi-DRIVE BUB1B and MAD2 dependencies, respectively; one-tailed t-test. (c) The sensitivity of near-euploid and highly-aneuploid cancer cell lines to the knockdown of BUB1B (top) and MAD2 (bottom) in the Achilles (left) and DRIVE (right) RNAi screens, across TP53 mutation classes. *, p<0.05; **, p<0.01, ***, p<0.001; two-tailed t-test. (d) The correlations between AS and the dependency on BUB1B (top) and MAD2 (bottom) in the Achilles (left) and DRIVE (right) RNAi screens, for cell lines that have not undergone whole-genome duplication (i.e., cell lines with basal ploidy of n=2). Spearman’s ρ = −0.32 (p=1e-05), −0.36 (p=7e-07), −0.30 (p=1e-04) and −0.28 (p=4e-04), respectively. (e) The sensitivity of near-euploid and highly-aneuploid cancer cell lines to the knockdown of BUB1B (top) and MAD2 (bottom) in the Achilles (left) and DRIVE (right) RNAi screens, after removing the effect of doubling time on gene dependency scores using linear regression. ****, p=1e-05 and p=9e-07, for RNAi-Achilles BUB1B and MAD2 dependencies, respectively; ****, p=1e-07; **, p=0.002; for RNAi-Achilles BUB1B and MAD2 dependencies, respectively; two-tailed t-test. (f) The sensitivity of near-euploid and highly-aneuploid cancer cell lines without microsatellite instability (MSS lines only) to the knockdown of BUB1B (top) and MAD2 (bottom) in the Achilles (left) and DRIVE (right) RNAi screens. ****, p=7e-07 and p=2e-07, for RNAi-Achilles BUB1B and MAD2 dependencies, respectively; ****, p=6e-07, for RNAi-DRIVE BUB1B dependency; ***, p=1e-04; two-tailed t-test. (g) The sensitivity of near-euploid and highly-aneuploid cancer cell lines to the knockdown of BUB1B (top) and MAD2 (bottom) in the Achilles (left) and DRIVE (right) RNAi screens, in cell lines that are WT for the 4 genes most selectively mutated in aneuploid human tumors (after TP53)12. **, p<0.01, ***, p<0.001; ****, p<1e-04; two-tailed t-test. (h) The sensitivity of near-euploid and highly-aneuploid cancer cell lines to the knockdown of BUB1B (top) and MAD2 (bottom) in the Achilles (left) and DRIVE (right) RNAi screens, after removing the effect of lineage subtype on gene dependency scores using linear regression. ***, p=4e-04; * p=0.015, for RNAi-Achilles BUB1B and MAD2 dependencies, respectively; **, p=0.002; * p=0.045, for RNAi-DRIVE BUB1B and MAD2 dependencies, respectively; one-tailed t-test. (i) The sensitivity of near-euploid and highly-aneuploid cancer cell lines to the knockdown of BUB1B (top two plots) and MAD2 (bottom two plots) in the Achilles (top) and DRIVE (bottom) RNAi screens, after removing the effect of HET70 scores on gene dependency scores using linear regression. ****, p=9e-07, p=8e-06 and p=5e-07 for RNAi-Achilles BUB1B, RNAi-Achilles MAD2 and RNAi-DRIVE BUB1B dependencies, respectively; **, p=0.001; two-tailed t-test.
