Figure 10.
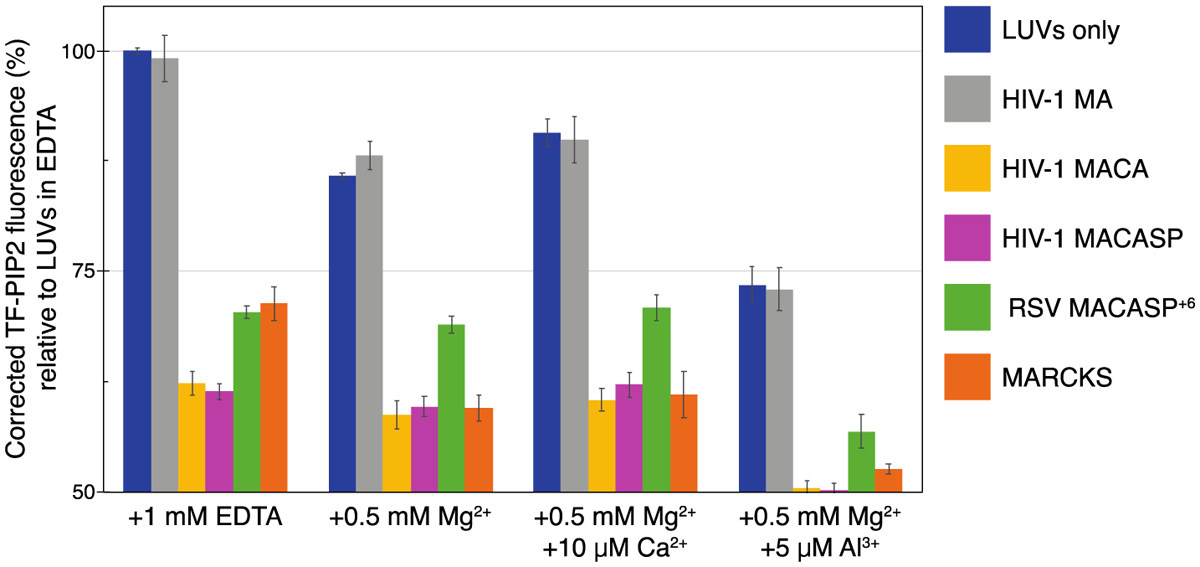
Proteins further promote PIP2 clustering of pre-existing multivalent cation-bridged PIP2. All LUVs were prepared in four different buffers that are based on 100 mM KCl, 20 mM HEPES, (pH 7.2), with additional EDTA or multivalent cations. PIP2 is free with 1 mM EDTA; PIP2 is modestly clustered with 0.5 mM Mg2+ or with 0.5 mM Mg2+ and 10 μM Ca2+, and PIP2 is strongly clustered with 0.5 mM Mg2+ and 5 μM Al3+. Note that lipids on both leaflets of the LUVs are exposed to the same buffer condition. Prior to protein addition, each protein/peptide was subjected to buffer exchange to match each buffer condition of LUVs. A total of 40 μl protein in each buffer was added to the outside of 160 μl LUVs in each buffer condition. TF-PIP2 fluorescence of LUVs mixed with buffer containing EDTA was maximum, set to 100%, while TF-PIP2 fluorescence of LUVs prepared with multivalent cations and/or mixed with protein or peptide was converted to the corresponding percentage. Note that for protein-induced PIP2 clustering, Y-axis is only shown from 50% to 100%. Each bar in the graph represents the average TF-PIP2 fluorescence % over the 1, 2, and 3 min time points post-mixing. Each quenching assay was performed at least three times; error bars show standard deviations from the means.
