Figure 2.
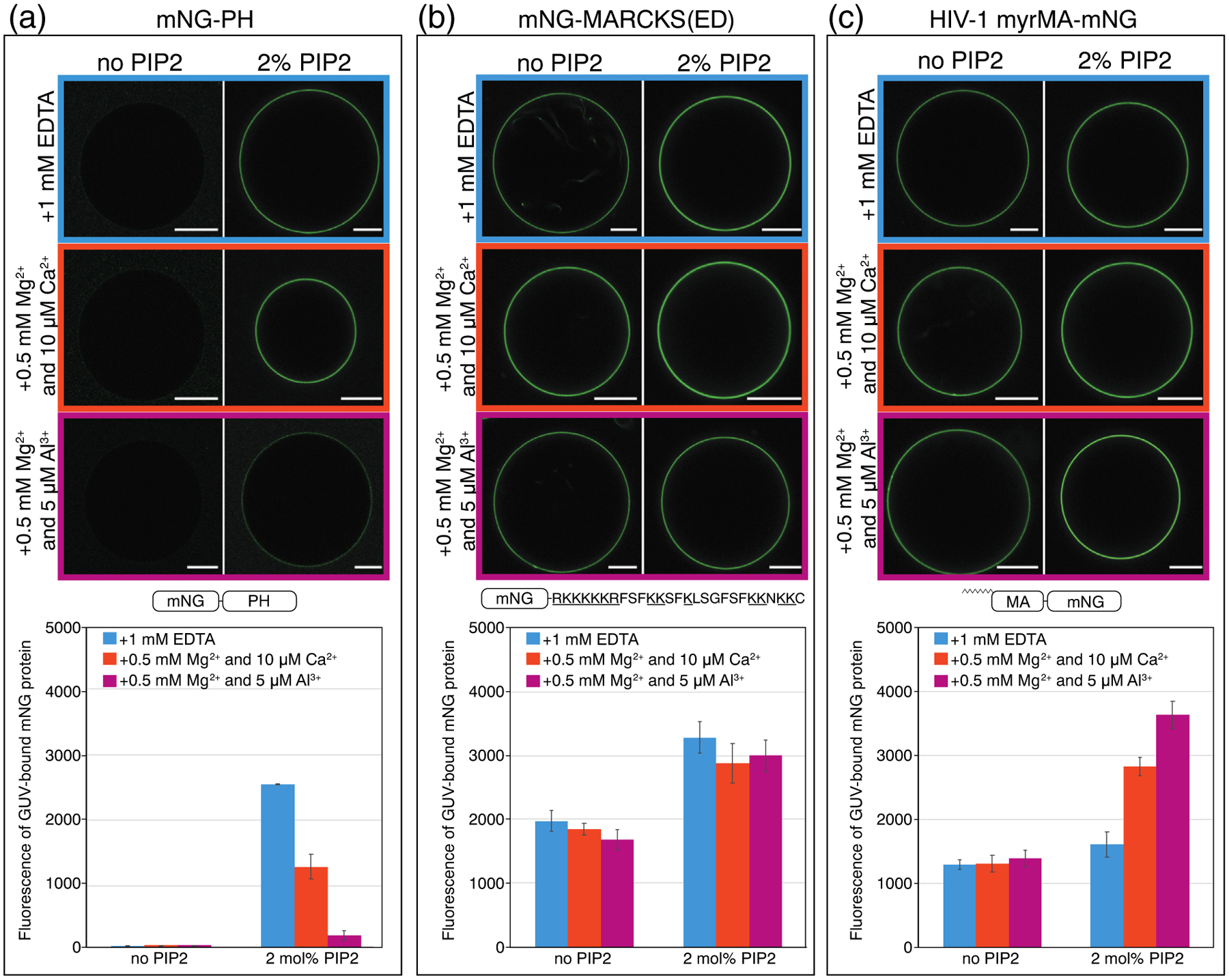
Myristoylated HIV-1 MA (myrMA) prefers binding to clustered PIP2, whereas the pleckstrin homology domain from phospholipase C (PH) favors binding to free PIP2. The myristoylated alanine-rich C kinase substrate effector domain (MARCKS(ED)) shows no preference. GUVs were prepared with no PIP2 [POPE/POPS/Chol (34/30/36)] or with 2 mol% PIP2 [POPE/POPS/Chol/brain-PIP2 (32/30/36/2)]. All GUVs were labeled with the membrane dye 18:1 DiI at a dye/lipid ratio of 1/2500. Both types of GUVs were harvested into one of three buffer conditions 5 h prior to protein binding. All three buffers were based on 100 mM KCl, 20 mM HEPES (pH 7.2), with either EDTA or multivalent cations. For GUVs incubated with 1 mM EDTA (top row), PIP2 is free; for GUVs incubated with 0.5 mM Mg2+ and 10 μM Ca2+ (middle row), PIP2 is modestly clustered; for GUVs incubated with 0.5 mM Mg2+ and 5 μM Al3+ (bottom row), PIP2 is strongly clustered. All proteins used in GUV assays were fused with mNG, with constructs shown in (a) mNG-PH, (b) mNG-MARCKS(ED), and(c) HIV-1 myrMA-mNG. Each mNG-labeled protein was added to the outside of GUVs at a final concentration of 1 μM. GUVs with protein bound were subject to confocal imaging under the same setting. Each GUV was examined at 488 nm for mNG fluorescence and at 561 nm for DiI fluorescence for locating GUVs. Fluorescence intensity above background (measured outside the GUV) at 488 nm (mNG) was analyzed by line scans in ImageJ across the perimeter of each GUV. The values were averaged and plotted with error bars representing the standard deviation. For each protein, a higher mNG fluorescence intensity indicates a higher membrane binding affinity. For GUVs with no PIP2, at least 15 GUVs were analyzed, and for GUVs with 2 mol% PIP2, at least 30 GUVs were analyzed, under each buffer condition. GUVs for each condition were prepared at least three times independently.
