Figure 6.
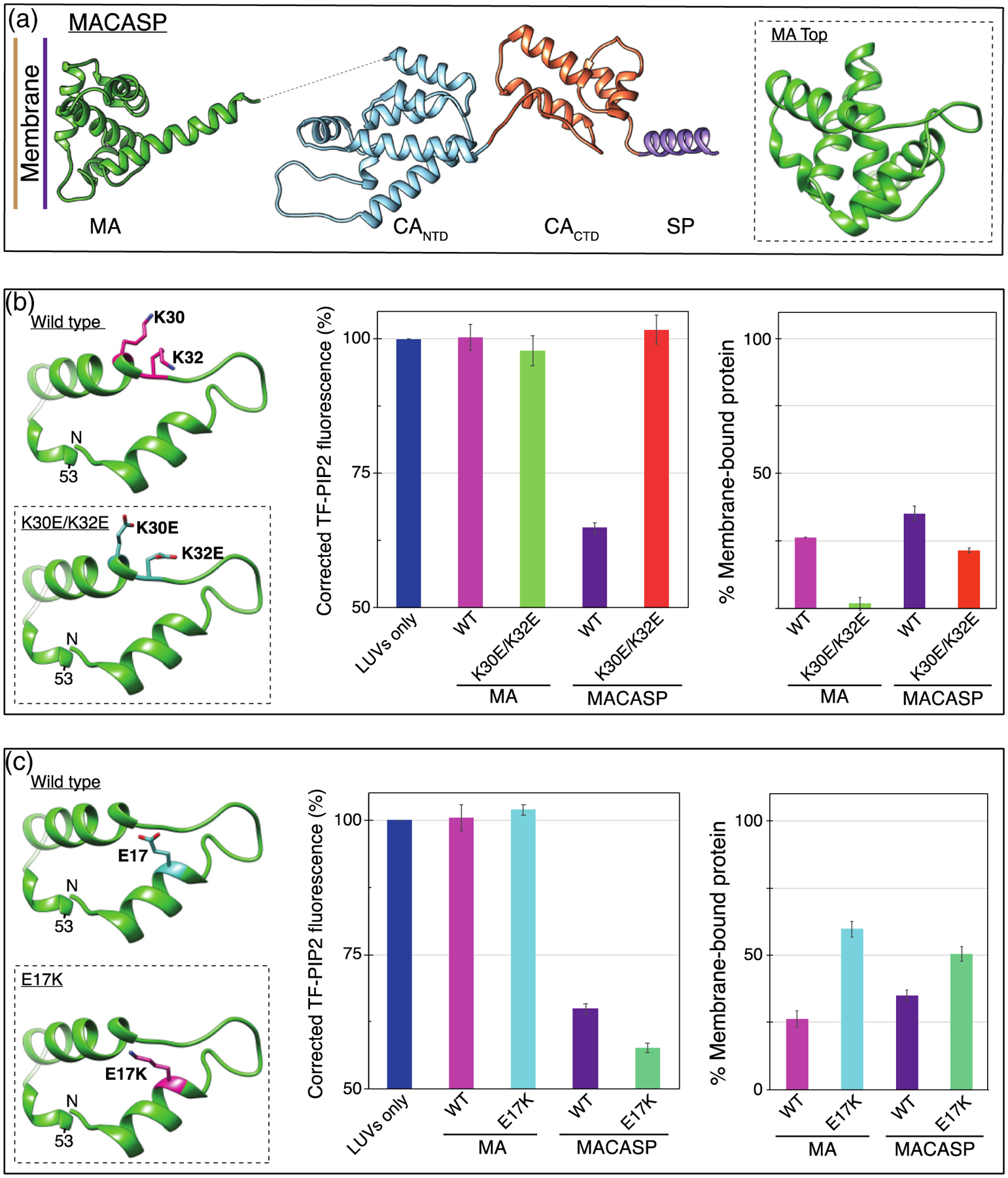
PIP2 clustering is dependent on known HIV MA PIP2-interacting amino acids. (a) Model of HIV-1 MACASP structure from PDB 1HIW (MA) [90] and 5L93 (CASP) [62]. Dotted line represents unstructured amino acids ~121–147. Inset is a top-down view of the membrane binding surface of MA. (b, left) MA membrane binding region, amino acids 7–53. PIP2-interacting amino acid K30 and K32 side-chains are shown in pink. Inset shows mutations K30E and K32E. (b, center) TF-PIP2 fluorescence in the absence and presence of wild-type (WT) or mutant HIV-1 MA or HIV-1 MACASP protein. (b, right) Membrane binding of each protein was determined by the pelleting assay. After incubating mixtures of 160μl LUVs and 40 μl proteins for 10 min, the protein-LUV mixture was ultracentrifuged at 75K for 15min at 4 °C. The supernatant was discarded and the pellet resuspended and subjected to SDS-PAGE and densitometry analyses. Each pelleting assay was performed at least three times with error bars of standard deviations from the means. (c, left) Amino acid E17 is shown in pink. Inset shows the membrane binding enhancement mutant E17K. (c, center) TF-PIP2 fluorescence in the absence and presence of WT or mutant HIV-1 MA or HIV-1 MACASP protein. (c, right) Membrane binding as described above.
