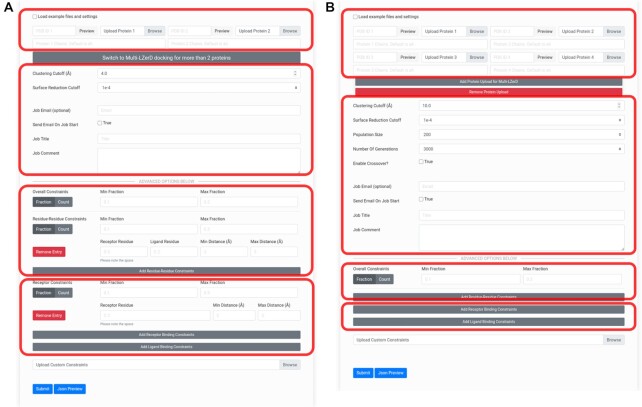
Figure 1.
Docking job submission interfaces. (A) The submission form for pairwise LZerD protein–protein docking jobs. The first area outlined in red is for specifying input subunit structures: the toggle allows users to quickly load example subunit structures and constraints, while the file upload buttons allow users to upload their own structures. Users can alternatively specify an entry to fetch directly from the PDB. Chain IDs to extract from the individual subunits can be entered. When a structure is loaded, a 3D interactive preview is displayed. Then, there is a button to switch to the Multi-LZerD form. The second area is for specifying general job parameters: the clustering RMSD cutoff controls the structural redundancy of the output models (higher RMSD means more diverse output structures), the surface reduction cutoff controls the fineness of the conformational sampling (lower means finer sampling), and optional user email and job title and comment settings control notifications and annotations. In the third and fourth areas, in the Advanced Options section, residue-residue or binding site constraints, respectively, can be added by clicking the Add buttons and filling in the residue number(s) and distance range fields. Users can toggle between min/max fraction and min/max count of constraints that must be satisfied. For more details, check the How to Use page on the server. (B) The submission form for Multi-LZerD multiple docking jobs involving 3–6 proteins. The Multi-LZerD form has Add and Remove buttons below the file upload buttons, which can be used to add or remove subunit structures. Any number of subunit structures from 3 to 6 may be uploaded. The areas outlined in red correspond to the outlined areas of the LZerD submission form.