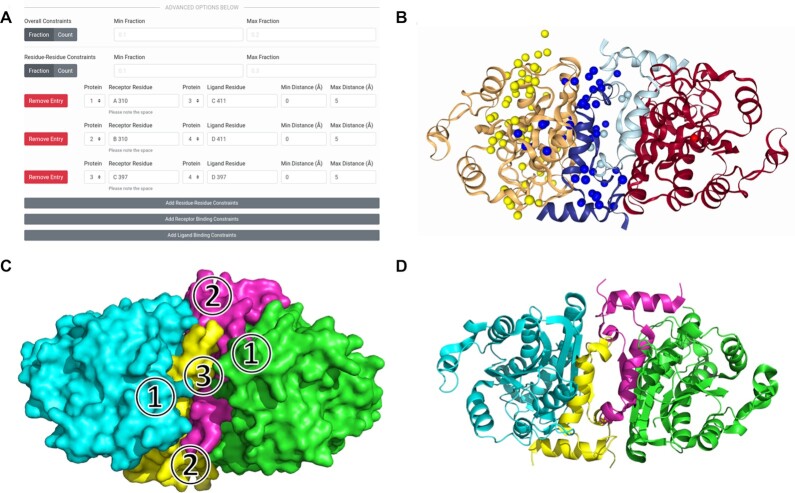
Figure 3.
Docking input and results for enoyl-ACP reductase multiple docking. This complex has four chains of size between 60 and 229 amino acids. (A) The constraints section of the Multi-LZerD submission form with the residue-residue interaction information to be integrated filled in. Here, distances in the range of 0 to 5 Å are specified for each of the pairs: subunit 1 chain A residue 310 and subunit 3 chain C residue 411, subunit 2 chain B residue 310 and subunit 4 chain D residue 411, and subunit 3 chain C residue 397 and subunit 4 chain D residue 397. The min/max fraction fields are blank, leaving the default that all constraints must be satisfied simultaneously. (B) The results of docking. The centroids of the top 50 models by ranksum are shown as spheres and are colored according to which subunit they represent. (C) A diagram of the protein-protein interfaces. In the top model shown in B, 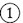 has an I-RMSD of 1.05 Å and an
has an I-RMSD of 1.05 Å and an 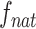 of 0.79,
of 0.79, 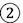 has an I-RMSD of 1.38 Å and an
has an I-RMSD of 1.38 Å and an 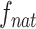 of 0.91, and
of 0.91, and  has an I-RMSD of 1.02 and an
has an I-RMSD of 1.02 and an 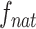 of 0.80. All three unique interfaces are modeled to medium quality under the CAPRI criteria. (D) The native structure of the complex (PDB 1NNU), shown superimposed to the same orientation as the docked models.
of 0.80. All three unique interfaces are modeled to medium quality under the CAPRI criteria. (D) The native structure of the complex (PDB 1NNU), shown superimposed to the same orientation as the docked models.