Figure 1.
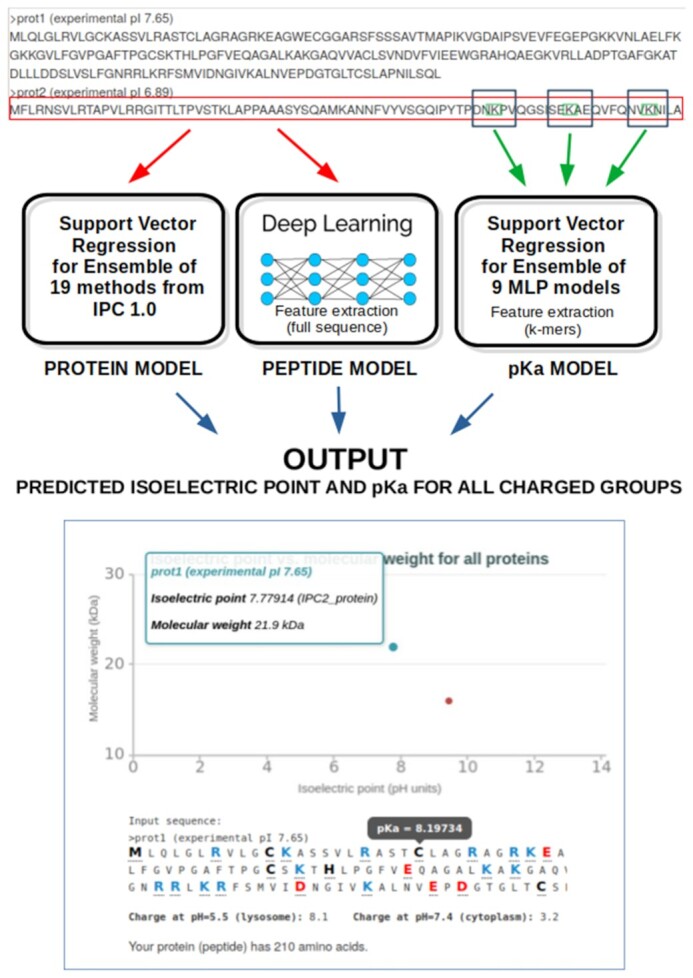
Overview of the IPC 2.0 architecture. The input (amino acid sequence in the plain format or multiple sequences in the FASTA format) is processed by individual machine learning models. Separate models depending the prediction task are used. Isoelectric point prediction for peptides is based on separable convolution model (four channels representing the one-hot-encoded sequence, AAindex features, amino acid counts, and predictions from IPC 1.0). The protein pI and pKa prediction models use the ensembles of low level models integrated with support vector regressor. For more details, see Supplementary Figure S1 and ‘Machine Learning Details’ in the Supplementary Material.
