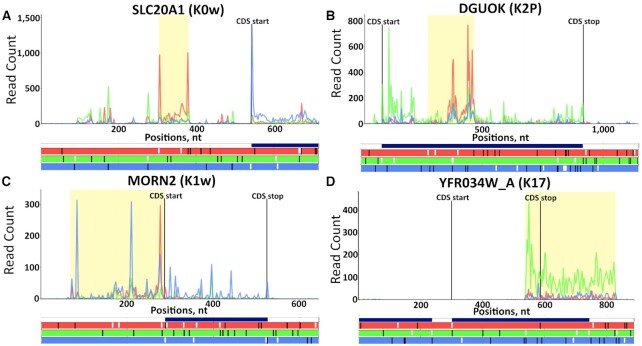
Figure 2.
Examples of highly ranked ORFs detected by Trips-Viz. At the bottom of each plot the open reading frame architecture is represented with three horizontal bars coloured red, green and blue to display each of the three reading frames. AUG codons are denoted by short white lines while stop codons are denoted by longer black lines. The CDS start and CDS stop positions are shown with the vertical black lines on the main plot along with counts of Ribo-Seq reads displayed in red, green and blue matching each of the three reading frames beneath. The ‘merged CDS’ bar above the reading frame bars displays a union of all annotated CDS regions in the corresponding locus. (A) An AUG-initiated uORF of the human gene SLC20A1 (Transcript ENST00000272542) in frame 1 (the annotated CDS is in frame 3). (B) A nested ORF in frame 1 of the human gene DGUOK (Transcript ENST00000264093) where the annotated CDS is in frame 2. (C) An N-terminal extension of the mouse gene Morn2 (Transcript ENSMUST00000061703). (D) An overlapping downstream ORF of the yeast gene YFR034W_A. Here the merged CDS bar extends into the 3′ trailer of YFR034W_A indicating the presence of another gene at the same locus. In this case the gene in question is YFR035C which is transcribed from the opposite strand, so it cannot explain the Ribo-Seq reads in the 3′ trailer of YFR034W_A. In addition, the reads extend beyond the merged CDS bar.