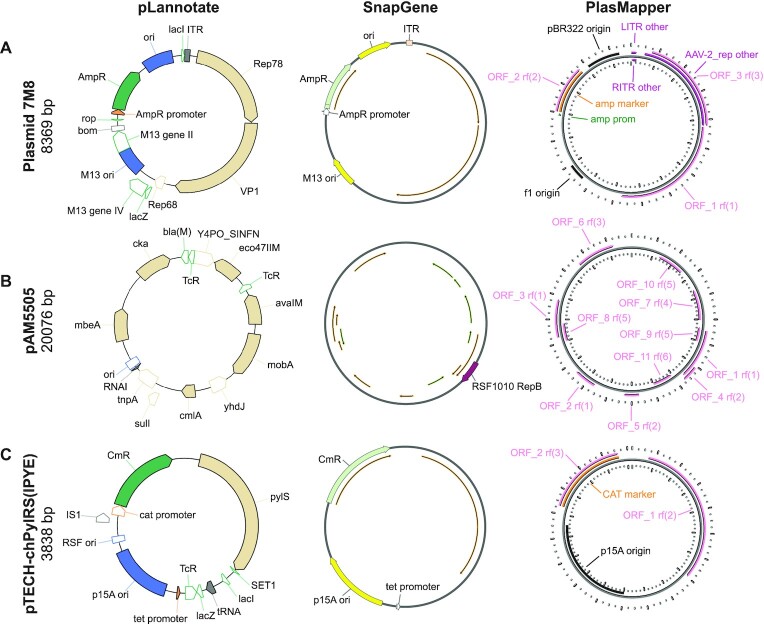
Figure 2.
Annotation results for three engineered plasmids. Each panel compares output from the pLannotate web server, the SnapGene viewer desktop program, and the PlasMapper web server. SnapGene and PlasMapper both have options to display generic open reading frames above a length cutoff that do not match specific features in their databases. These ORFs are displayed as thin arrows for SnapGene results and pink arrows for PlasMapper results. pLannotate displays features in different colors based on their type if they are matches to the GenoLIB feature database and in tan if they are matches to proteins in Swiss-Prot. Fragmentary matches are shown as unfilled arrows. Labels were manually edited to improve legibility for all tools. (A) Plasmid 7M8 is used for adeno-associated virus (AAV) vector production. (B) Plasmid pAM5505 is a helper plasmid for engineering cyanobacteria via conjugal transfer of DNA. (C) Plasmid pTECH-chPyIRS(IPYE) contains an engineered aminoacyl-tRNA synthetase enzyme. Note the partial TcR open-reading frame downstream of the tet promoter that is discussed in the text