Abstract
Advances in two-dimensional (2D) and three-dimensional (3D) cell culture over the last 10 years have led to the development of a plethora of methods for cultivating tumor models. More recently, cellular co-cultures have become a suitable testbed. The first portion of this review focuses on co-culturing methods that have been developed in recent years utilizing the multicellular tumor spheroid model. The latter portion describes techniques that are used to analyze the proteomes of mono- or co-cultured tumor models, with a focus on mass spectrometry (MS)-based analyses. Protein profiles are important indicators of the tumor heterogeneity. Therefore, there is a specific focus within this review on analysis by MS and MS imaging methods evaluating the proteomic profiles of 2D and 3D co-cultures. While these models are incredibly important for biological research, so far, they have not been widely explored on the proteomic level. With this review, we aim to introduce these systems to an analytical audience, with the goal of highlighting MS as an underutilized tool for proteomic analysis of tumor models.
Keywords: cancer, co-culture, mass spectrometry, spheroids, tumor microenvironment
1 |. INTRODUCTION
Cancer is a complex disease that can be triggered by acquired or inherited genetic changes in cells producing a clonal population of abnormal epithelial cells, referred to as a neoplastic population. These neoplastic cells are the foundation of cancer, as they initiate malignant progression, while spreading oncogenic and tumor suppressor mutations throughout tissue as they multiply [1]. The chemical and physical complexity within tumors has caused a shift in the cancer community in how the disease is studied from both the molecular and phenotypic viewpoints. Previously, tumors were viewed simply as masses of proliferating cells, however, in recent decades, that paradigm has evolved to consider combinations of cell types and non-cellular components that interact and communicate with each other. The tumor is now rarely considered in isolation but in concert with a collection of variables that all contribute to the surrounding tumor microenvironment (TME).
The TME contains several cellular components including epithelial, endothelial, stem, and immune cells, any of which have the potential to develop into a neoplastic cell. The neoplastic, or cancerous, cell population carries genetic changes that convey replicative immortality and sustained proliferative signaling to maintain control [1]. These cells proliferate at a higher rate than normal cells and contain perturbed protein profiles. Both of these qualities can help cancer commandeer control of the cell cycle. Within the cancer genome, mutations such as those activating oncogenes, or inactivating tumor suppressor genes, can benefit or hinder tumor progression. The tumor protein 53 (TP53) gene is commonly mutated, which benefits tumor progression across a large number of cancer types [2]. The Cancer Genome Atlas Pan-Cancer project has found these mutations among 3,200 cancer patients with 12 tumor types. The mutations vary in location on the gene; however a majority of the mutations result in the impairment of the p53 pathways both identified and unidentified [3]. The prevalence of this mutation across several cancers has made it a popular target for drug therapy. Mutations to the TP53 gene and a plethora of other known and unknown mutations lead to uncontrollable replication of neoplastic cells, and the subsequent reprogramming of non-neoplastic components present in tissue.
Cancer cells must recruit other stromal elements, as well as molecules from the extracellular matrix (ECM) in the surrounding environment for their own survival [4]. This recruitment can occur through a variety of juxtracrine or paracrine signaling mechanisms. For example, hypoxia in the TME can spur angiogenesis. Hypoxic conditions elicit cancer cells to secrete soluble factors such as vascular endothelial growth factor A (VEGFA) [5]. VEGFA as well as other pro-angiogenic mediators induce the activation of tip cells, or motile endothelial cells. Tip cells degrade the surrounding ECM and migrate to form new vascular sprouts towards the chemokine [6]. Cancer-associated fibroblast (CAF) cells have been found to respond to soluble factors secreted by tumor cells VEGFA or TGF-B to contribute to angiogenesis and remodel the surrounding ECM through matrix metalloproteinases and increased ECM protein secretion respectively [7]. The components within the TME work together to not only maintain control of local tissues, but also to trigger metastasis and invasion, spreading to other portions of the body.
With the considerable complexities of cancer, the models that are used to study the disease must also reflect the multifarious TME components. Models are useful in preliminary research for studying drug interactions with cancer cells before clinical trials, as well as obtaining information about the changes in the biochemistry and phenotype of TME cells. The simplest tumor model developed is the two-dimensional (2D) monolayer culture of epithelial cancer cells [8]. Models have since been established to be more complicated and better represent the TME. Three-dimensional (3D) tumor models are considered to better represent the TME as compared to 2D models. In addition, as co-cultures are more beneficial than mono-cultures in simulating in vivo conditions, the majority of work that utilize tumor models have used monoclonal models [9]. Within the past decade, there has been a push in the field towards co-culturing models to include multiple elements of the TME.
Co-culturing multiple cell lines into a single 3D model approximates elements intrinsic to in vivo conditions such as paracrine signaling, cell-to-cell communication, and modeling various phenomena such as the epithelial to mesenchymal transition (EMT) [10]. Modeling the TME can be approached as building a pyramid. At the base of the pyramid is a neoplastic, monoclonal culture, which is often an epithelial cell line, or patient-derived tissue. As the pyramid is built up from the base, additional TME components are recruited by the neoplastic population to help provide infrastructure, protection, and aid in malignant progression. These additions can be one or more non-neoplastic elements, such as fibroblasts or immune cells. Intra- and intercellular interactions along with protein alterations within the cell can be monitored, utilizing various methods such as mass spectrometry (MS), fluorescence, and other proteomic techniques that will be discussed later in this review.
Previous well written review articles have described methods of culturing 2D and 3D monoclonal models [11–13]; however, there is a lack of centralized information regarding co-culturing cancer models. There is an underutilization of co-culturing techniques in the proteomics realm, and a dearth of studies investigating 3D cellular models with MS-based analysis techniques. MS is a diverse technique that has been shown to be compatible with 3D cultures and would have great promise in analyzing novel co-cultures.
2 |. VARIABLES WITHIN TUMOR MODELS: 2D VERSUS 3D TUMOR MODELS
2D cell culture as a model for the TME is limited in the information that can be obtained. This simple model has helped develop an understanding of complex cellular physiological functions and pathways, as well as how cells respond to various stimuli [13]. Due to the coplanar nature of the model, many aspects of the TME cannot be recapitulated. The gene and protein expression profiles in 2D cultures do not emulate modulations that occur in vivo, which indicates that proteomic studies of 2D cultures could be inaccurate [12]. This model is easy to generate, which makes it a popular choice for routine studies.
There are greater varieties for 3D culture models regarding culturing methods [9,12,14,15]. Nevertheless, 3D tumor models share the common characteristic of better representing the complexity of in vivo TME conditions, including the gene and protein expression profiles [16]. Multicellular tumor spheroids (MCTS), often referred to as spheroids, are widely used tumor models for proteomic studies of the TME. In comparison to 2D models, MCTS have increased complexity in their structure to create nutrient and oxygen gradients for closer replication of in vivo conditions [17]. MCTS are formed by seeding cells suspended in media into an environment where they are unable to adhere to a surface. By preventing attachment to a 2D surface, cell to cell attachment is encouraged, and the resulting product is a spherical shaped tumor model. Through this growth pattern, MCTS can recreate distinct zones within a tumor (Figure 1). In addition, they can recapitulate glucose flux rate, hypoxia, display cancer stem cell (CSC) subpopulations, and glucose flux rate of in vivo tumors [18]. MCTS have a high reproducibility, are compatible with high-throughput studies, and are considered to be easy to cultivate [19].
FIGURE 1.
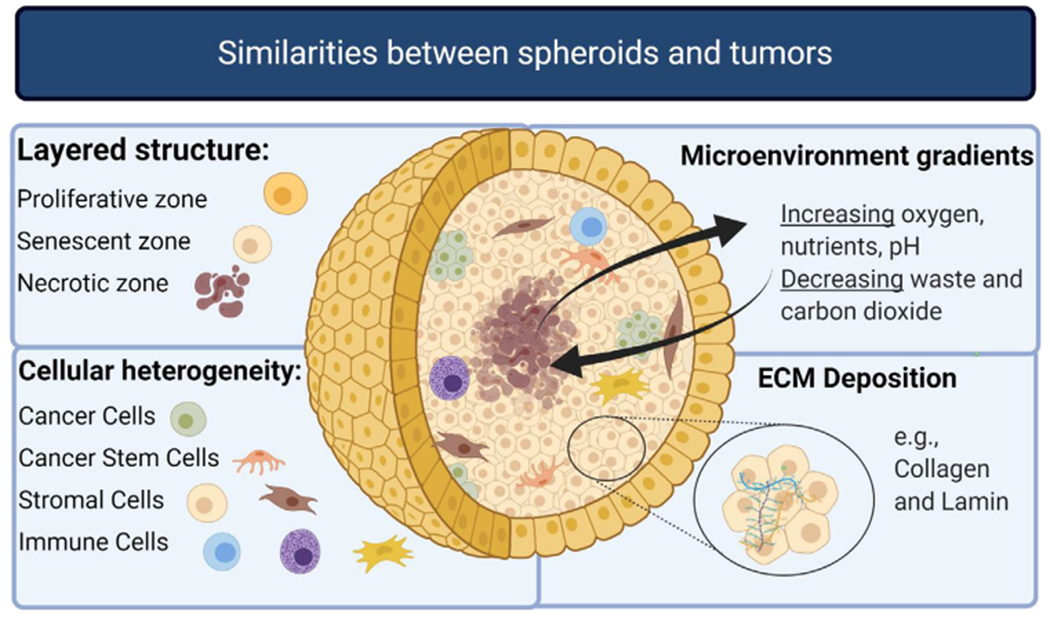
Spheroids emulate aspects of the TME such as the layered structure, the tumor heterogeneity, various microenvironment gradients, and ECM deposition. This figure is adapted from Costa et al. [18] Made with biorender.com
2.1 |. Culture methods: Scaffold versus scaffold free
As monoclonal MCTS can recapitulate the solid structure of an avascularized tumor, co-culturing MCTS allows for further complexities of the TME to be analyzed. Interactions of neoplastic and non-neoplastic populations cannot be captured within monocultures; however co-culture offers the ability to study cancer biology that stems from these interactions. MCTS co-culturing methods use similar methods of cultivation, with small deviations. The additives to the media or the ratio of cellular components that are initially seeded into the model may vary upon optimization. Numerous methods of cultivating MCTS have attempted to capture the complexity of tumor and genomic heterogeneity. These cultivation methods can usually be placed into one of two categories, those that do and do not use scaffolds for infrastructure.
Scaffolds serve the purpose of simulating certain aspects of the ECM in vitro to aid tissue growth. Scaffolds fall into one of the three classes described as biological, synthetic, or a hybrid of both. Their definition is reliant on the material they are made from and the properties they possess. Culture methods that use scaffolds or matrices provide the cells with a biologically active environment by forming microstructures that cells adhere to in interstitial spaces. Using a scaffold can be beneficial if creating a 3D model utilizing a cell line that does not naturally aggregate into a spherical shape [20]. More information regarding the procedural use of scaffolds can be found in reviews focused predominately on culture methods [21–23]. Scaffolds enable faster remodeling to the tissue, ECM decomposition, and allow for the culture to be implanted into a host tissue more easily. Unfortunately, these platforms have low reproducibility stemming from inevitable lot to lot variation amongst matrices [24]. Other issues arise when therapeutics adsorb onto the surface of the scaffold, resulting in uneven dosing, or contaminants and residual growth factors that are introduced when using animal-based scaffolds. Additionally, all scaffolds are not compatible with imaging analysis methods based on the transparency of the material from which they are formed, due to the light scattering requirement for imaging techniques. Certain scaffolds do not possess the ability to scatter light, and therefore cannot be compatible with imaging [25].
Synthetic scaffolds are polymers that take on a less active role in cultivation and are merely physical supports for cells [26]. Hydrogels are water-insoluble polymers that contain tissue-like elasticity and can retain a high capacity of water due to micropores. Such micropores support the surrounding tissue in facilitating the transport of oxygen, nutrients, growth factors, and metabolic wastes. This transport mimics how similar particles would move through an in vivo TME. Matrigel is a widely used, ECM-based natural hydrogel formed from secreted basement membrane extracts of mouse sarcoma cells, allowing for a scaffold rich in ECM components and factors to be used by cells growing around the Matrigel matrix [27].
Liquid overlay is the most prevalent scaffold-free technique for producing MCTS (Figure 2). Spheroids are cultivated in a round bottom well where the cells cannot physically attach to the surface, for example, an ultralow attachment plate coated in polystyrene, or a well-plate with agarose at the bottom to block cell attachment. The seeded cells aggregate in each well to form a central nexus, then grow outward in a radially symmetric fashion to form the spheroid shape. This method is a popular choice as it is easy to culture, relatively inexpensive, and allows easy access to each individual spheroid for drug dosing and growth monitoring. In addition, the MCTS produced are homogenous in shape and size [28]. Certain biomaterials can create challenges for coating the vessel surface. For example, poly-hydroxyethyl methacrylate is complicated in its preparation and use, although using agarose will avoid this issue [28].
FIGURE 2.
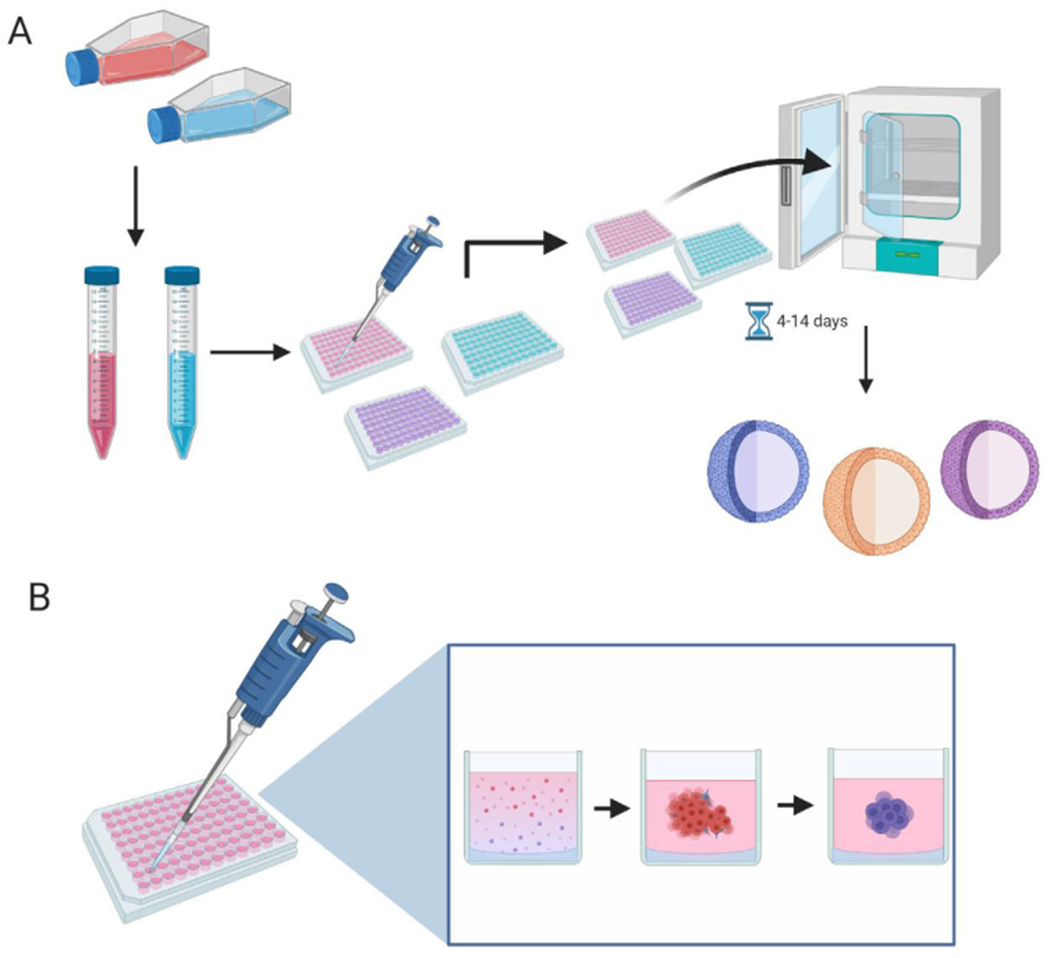
Methodology of the liquid overlay technique used to generate MCTS. (A) Depicts confluent adherent cells trypsinization, and subsequent seedining into ultra-low attachment or agarose laden 96-well paltes. The plates are then cultured for 4–14 days, with media changes if necessary. (B) Depicts the process of how a MCTS forms within a single well. The low attachment conditions promote cell to cell adhesion, resulting in the cells forming a spheroid shape. Made with biorender.com
2.2 |. TME cellular components
The TME is a complex system of neoplastic and non-neoplastic cells, ECM, structural vessels, and signaling molecules. These components modulate the TME in pathways that contribute to the survival and growth of a tumor by communicating via cells to cells, stroma, or ECM interactions [29]. Examples of studies previously conducted with co-culture and the corresponding TME components used are detailed in Table 1. Epithelial cells are the origin of neoplastic populations in organ-based cancers such as skin, breast, colon, lung, pancreatic, and urinary/genital cancers, due to the large turnover of epithelial cells [30].
TABLE 1.
Examples of co-culture models that have been used and their analysis techniques
| 3D model used | Components used | Disease and cell lines used | Analysis techniques | Conclusions | Reference |
|---|---|---|---|---|---|
| Tumor on a chip with microfluidics | Epithelial (adenocarcinoma) and endothelial | Breast cancer (MCF-10A, MCF-7, MDA-MB-231); human umbilical vein endothelial cells | Lumen microscopy, fluorescent staining | Established microfluidic breast cancer model for endothelial-epithelial crosstalk | Devadas et al. |
| Tumor on a chip with microfluidics | Epithelial and fibroblast | Prostate epithelial (iPrEC, EMP); human stromal fibroblasts (BHPrS1) | Immunofluorescence microscopy | PIM1 induction observed to convert stroma to CAFs supports prostate-on-a-chip model | Ivich et al. |
| Spheroids | Epithelial and fibroblasts | Colon cancer MCTS (HT-29, DLD-1); colon and lung fibroblasts (CCD-18co, WI-38) xenograft control) | Immunohistochemistry, ELISA, Western blot, RT-qPCR, human cytokine array analysis, APH dose-response assay | Increased EMT-related protein expressions | Kim et al. |
| Spheroids | Epithelial and fibroblasts | Colon cancer MCTSs (HT116, Ht-29); colon fibroblasts (CCD-18Co) | Genomics screening (shRNA), ELISAs, RNAseq | Identified CAF derived protein targets for drug discovery | Horman et al. |
| Spheroids | Epithelial and stromal | Human endometrial cancer (IK); human endometrial stromal cells | Histological and immunohistochemical analysis (IF), LC-MS/MS | 591 Common proteins signifying phenotypic similarity to in vivo endometrial tumors | Al-Juboori et al. |
| Spheroids | Epithelial and stem | Ovarian cancer (CAOV3, A2780, SKOV3); Met-5A mesothelium cells | Immunohistochemical staining, qRT-PCR | Facilitation of spheroid formation and adoption of cancer stem-like characteristics | Shishido et al. |
| 2D microfluidics | Epithelial and endothelial | Cervical cancer (CaSki); human umbilical vein endothelial cells | Flow cytometry, ESI-MS | Integrated microfluidic model of tumor-endothelial interactions for drug screening | Lin et al. |
| Spheroids | Epithelial, stem, fibroblasts | Colon cancer MCTS (SW620); mesenchymal stem-like (3A6), foreskin fibroblasts (Hs68) | Immune-fluorescence staining, Western blot | Enhancement of cytotoxicity resistance from chemotherapy | Tsai et al. |
| Spheroids | Epithelial, immune, and fibroblasts | Colon cancer (HT-29); monocytic cells (THP-1) and fibroblasts (175BR) | Activity assay, Western blot, RT-PCR, siRNA gene silencing | Elevated invasive potential correlated to cathepsin B expression | Krueger et al. |
| Spheroids | Epithelial, fibroblasts, endothelial | Pancreatic PANC-1 MCTSs; human lung fibroblasts (MRC-5) and HUVEC | Histological analyses and selective plain illumination microscopy | Scaffold free MCTS model with multiple stoma to mimic drug resistance | Lazzari et al. |
| Spheroids | Epithelial, immune, and fibroblasts | Colon cancer MCTS (HCT-116); human intestinal fibroblasts and monocytes | Immunohistochemistry, flow cytometry | Nanoparticle uptake comparison in mono, double and triple culture | Baulth-Ramos et al. |
| Spheroids | Epithelial and fibroblasts | Colon cancer (DLD1) NIH3T3 fibroblasts | Immunofluorescence, qPCR | Fibroblasts display different behavior in 3D and 3D co-cultures and functional gene analysis using automation compatible 3D cell culture models have the potential to discover novel target genes or drug development | Thoma et al. |
Epithelial cells within normal tissue provide protection by forming sheets to cover the skin and the walls and channels of cavities within organs [31,32]. Multiple epithelia-containing tissues are similarly structured and result in the most common human cancers. In cancerous tissue, signals from cancer-associated stroma can trigger the expression of EMT transcription factors, changing the cells into the spindle-shaped mesenchymal morphology [33]. Certain breast and colorectal cancers (CRCs) are hypothesized to originate through the EMT, making it a relevant process to study within the context of cancer [29,34,35].
Epithelial and mesenchymal cells express different molecular markers, which are dependent on the EMT. The EMT phenomena are described as a sliding scale of discrete states, with cells presenting a phenotype closer to epithelial or mesenchymal cells. This makes it difficult for epithelial cells to be identified in functional studies, and has prompted researchers to assign epithelial cells an EMT score [36]. Due to this property, a cell that is epithelial in origin, but displaying a phenotype closer to a mesenchymal state could fail to express traditional epithelial markers while still expressing markers considered to be mesenchymal [33]. Furthermore, many mesenchymal cells express molecular markers that overlap with those expressed by fibroblasts.
Fibroblasts are spindle shaped cells with a flat, oval nucleus at rest, and display a stellate morphology when activated. In normal tissue, fibroblasts are activated during injury, and generate growth factors, lysl oxidases, ECM proteins such as collagens and fibronectin, and matrix metalloproteinases (MMP) for reconstruction of the tissue [37,38]. The function of normal fibroblasts can be influenced by neoplastic cells through signaling, leading to the formation of CAFs. CAFs predominately encourage tumorigenesis by promoting an immunosuppressive, inflammatory, oxygen-rich microenvironment through the excretion of pro-angiogenic (e.g., VEGFR) and myofibroblast (e.g., vimentin, desmin, FSP1) signals [37,39].
CAFs are difficult to study as they have the ability to differentiate into distinct subpopulations. Additionally, they express molecular markers that are also expressed by other cells within the TME, including mesenchymal, endothelial, and immune cells [40,41]. Within the field of translational cancer research, there has been a large amount of difficulty identifying one specific marker for fibroblasts. The overlap in expression of multiple cellular populations tends to be heterogenous within cancer types [42,43]. It has become commonplace to utilize at least two or more fibroblast specific markers when trying to characterize these cells. The prevalence of subpopulations of fibroblasts makes it difficult to fully study the intricacies of fibroblasts within the TME [41].
Endothelial cells are non-neoplastic cells that are not initially a component within the microenvironment but can be recruited by tumor cells to spur the formation of new blood vessels from existing vascular structures. The conscription of endothelial cells through pro-angiogenic factors (e.g., cytokines, growth factors, ECM proteins, ECM remodeling enzymes, extracellular vesicles [EVs]) results in perfuse blood vessels to the TME. This is due to a lack of basement membrane in tumor vasculature, and the poor quality of blood vessels formed often creates hypoxic conditions within the TME [5]. Once recruited, endothelial cells are referred to as tumor-associated endothelial cells. These cells have several genetic expression signatures that distinguish them from normal endothelial cells, such as elevated VEGF receptor protein expression. The upregulation of the VEGF receptor tyrosine kinase enables the VEGFR signaling pathway to mediate survival, vascular permeability, migration, and proliferation through downstream signaling [44].
CSCs are unique cells and are defined by the ability to self-renew and beget heterogenous lineages of cancer cells that compose the tumor. In normal tissue, stem cells serve the main function of differentiation for allowing organs and tissues to maintain their functions through a lifetime [45]. CSCs are not necessarily derived from normal adult stem cells, and they are not composed of embryonic stem cells. CSCs do share some stem cell transcriptional programs with both embryonic and CSCs and take cues from niche environments [46].
As CSCs are composed of various heterogenous niches that may express different markers, they are identified through functional validation from well-established assays. These include cell culture assays (tumor initiating assays and self-renewal assay), or through the identification of molecular markers specific to the type of cancer in which the CSC originated. The need to functionally validate the self-renewal capabilities of a CSC makes this a difficult component to study. Additionally, molecular markers for CSC may change over time in response to TME conditions or cell cycle-related expression. Many of the molecular CSC markers also use their utility when cells are cultured in vitro [47].
Evading immune destruction is an emerging hallmark of cancer, which is reflected by a severe increase in research regarding the immune response to cancer [1]. This review will not go in depth to explain the intricacies of the immune response to cancer, as it deserves its own separate article [48]. Generally, immune cells sent to the tumor site are referred to as infiltrating immune cells. Early infiltrating cells include macrophages, lymphocytes, natural killer cells, and dendritic cells. These cells have duality in the TME, as they can contribute towards tumor immunity through the release of growth factors (chemokines, interleukins) or release factors that can aid in forming a tumor suppressive environment (B cells, Treg, tumor-associated macrophages) [49]. In recent years, immunotherapy, using the immune system of individuals to help fight cancer, has gained prevalence as a treatment. Within immunotherapy, specificity to cancerous cells is generally increased and leads to better patient outcomes. Although, it has been found that cancer can develop mechanisms of escaping the immune response.
2.3 |. ECM
The ECM in normal tissue is comprised of a 3D network of various macromolecules, including integrins, collagens, proteoglycans/glycosaminoglycans, elastin, fibronectin, laminin, as well as other glycoproteins [50]. The ECM is perhaps the most important component of the TME, as it generates specific bidirectional communication between cells. This communication contributes to the mechanisms for cell fate and the behavior of cancer cells [51]. Cells within the TME communicate by specific ligand-receptor interactions. For example, E-cadherin, the transmembrane protein that mediates homophilic cell–cell interactions, communicates with integrin, which communicates with the ECM. E-cadherin mediates Ca2+-dependent homophilic interactions with opposing molecules in neighboring cells and acts as a tumor suppressor [52].
The ECM in normal tissue has self-remodeling capabilities. The ECM in cancerous tissue retains this function, utilizes this ability to form a natural scaffold for tumors, and can use this remodeling to enhance communication. Cells sense binding motifs of ECM proteins and integrin binding events on the cell surface and restructure their surroundings to allow for signaling molecules to move through the space to communicate [9]. During metastasis, cells travel through the remodeled matrix [14]. Construction of the matrix occurs through deposition of ECM proteins by TME cell components and degradation by a family of cleaving proteins called metalloproteinases(MMP). Both processes are regulated by integrin signaling pathways and are controlled largely by the secretion of soluble factors by fibroblasts.
3 |. MS-BASED PROTEOMICS FOR SOLVING BIOLOGICAL SYSTEMS
The term “proteome” was coined at a conference in Siena, Italy in 1994. Since this time, the world of proteomics has matured into the versatile, highly sensitive methodology that it is today [53]. Prior to this conference, hard ionization methods had made it difficult to analyze nonvolatile, biological compounds. The development of highly sensitive ionization methods for complex biological molecules, such as proteins and peptides, aided proteomics in gaining traction in the scientific community. Throughout the late 20th century and the early portion of this century, methods using electrospray (ESI) and matrix-assisted laser desorption/ionization (MALDI) were introduced and refined. ESI was found to couple well with triple quadrupole mass or ion trap mass analyzers. MALDI sources were often paired with time of flight due to its ability to separate larger ions through the flight path of the TOF.
Older gel-based methods to analyze protein expression utilize a forward (function to sequence) approach, or a reverse approach where the protein profiles expressed within cells or tissues are compared. 2D electrophoresis methods separate proteins based on specific qualities with high reproducibility and with the ability to quantify the protein [54]. Patterson and Aebersold wrote a comprehensive review outlining the developments that formed proteomics as a field [53]. Within the review, they outline how proteomics are beneficial to study biological systems using a systems biology approach. The systems biology approach can also be applied to understanding the TME. Studying the TME with co-cultured tumor models and MS allows for a researcher to obtain a global snapshot of the proteins present at the time of analysis. These proteins can be identified/quantified to understand cellular processes that drive malignant progression.
Currently, multiple analysis methods are used tangentially to assess co-cultured tumor models on a proteomic level; these methods can be divided into MS-based and non-MS-based methods. Non-MS methods include the use of antibodies for visualization of specific proteins using immunohistochemistry or immunofluorescent imaging, PCR-based methods of protein separation, and assays to measure numerous biological properties. When used individually, these methods lack the ability to fully capture the minute perturbations that co-culturing can produce in the TME. MS-based methods for proteomic studies typically use ESI-MS/MS, and MALDI-TOF. These ionization methods and corresponding analyzers are capable of elucidating how variations in conditions, such as the addition of a TME component, modulates the environment through analyzing each peptide or protein that is able to be isolated, fragmented, and detected [35]. High-throughput proteomics in combination with MS serve as a platform to characterize thousands of proteins within the complex TME, and should be used to further identify the subtle changes in co-culturing tumor models.
Traditional proteomics approaches are top-down or bottom-up (Figure 3), although specifically tailored approaches that include middle-down are gaining in popularity. Top-down proteomics separates complex protein mixtures within the mass spectrometer and analyzes proteoforms within the complex mixture. Top-down studies are therefore useful in analyzing different forms of the same protein that have been modified through post translational modifications, alternative splicing, or genetic variation. Bottom-up proteomics involves protein separation before the protein mixture is analyzed with the mass spectrometer. This analysis requires digesting proteins into peptides and separating these peptides through methods such as reverse phase liquid chromatography, small cation exchange, or metal affinity chromatography [25]. The majority of MS-based methods in Table 1 employ bottom-up proteomics. Cancer research that employs proteomics can be split into four sections by the application of the research: biomarker discovery, prediction of chemotherapy responses, biology of malignant progression, and database contributions.
FIGURE 3.
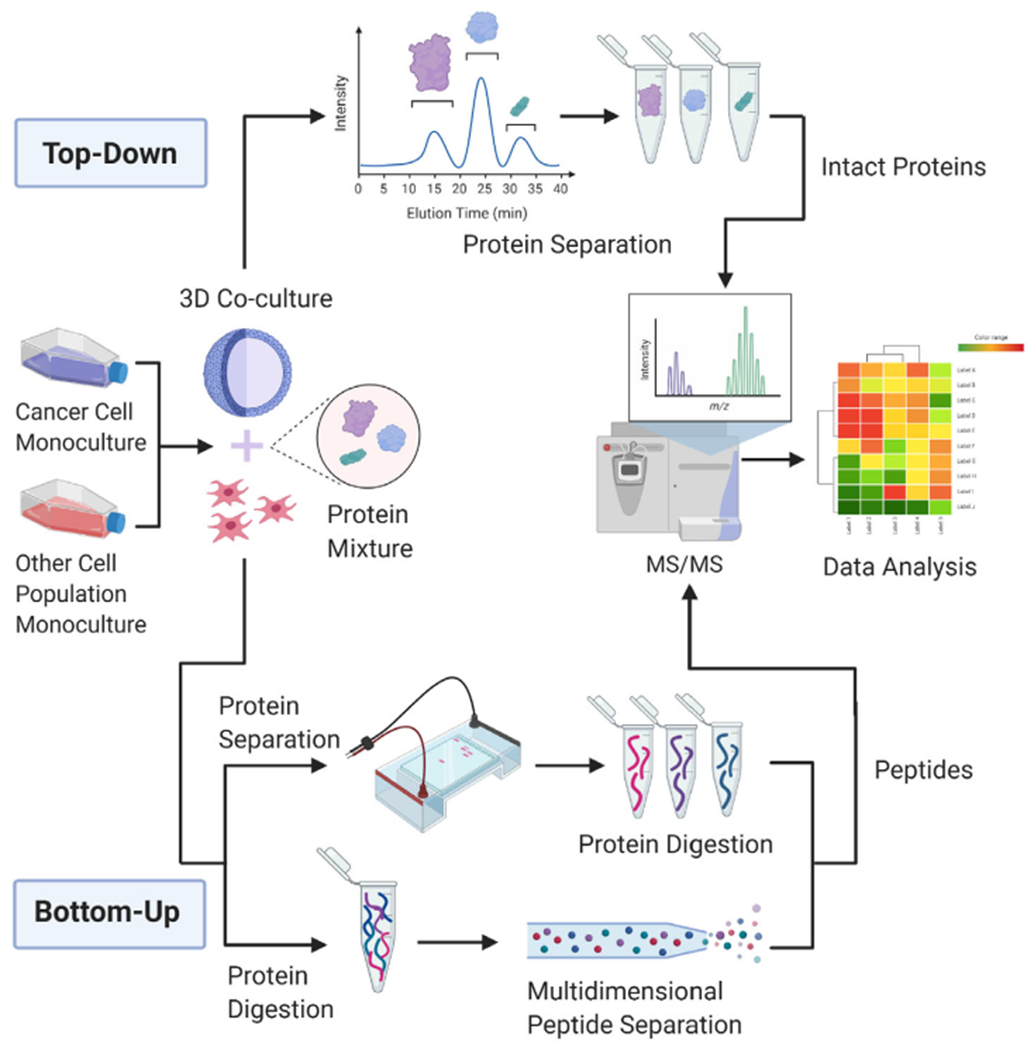
A schematic detailing top-down and bottom-up proteomic methodologies. Two populations of cells are cultivated to confluency, then seeded into non-adherent conditions to form spheroids. After the cultivation period, spheroids are harvested and proteins are extracted. Top-down requires the separation of intact proteins through LC-MS/MS. Bottom-up proteomics requires further sample prep before running a peptide solution through the mass spectrometer
3.1 |. Prediction of chemotherapy responses
Proteomics can not only capture the full proteome of a cell, but the perturbed protein profile of a cell with the same genotype that has been treated with a drug. A proteomic approach permits for the protein-based perturbations of the TME to be analyzed to indicate which cellular processes are affected by the addition of a drug. This has previously been extensively performed using monocultures, and in recent years co-cultures have been used to help further understand the TME. With the implementation of multiple components in co-cultured tumor models, bottom-up quantitative proteomics analysis methods have aided in the understanding of how the TME will respond to chemotherapeutic drugs and should be used with more frequency.
Older quantitative proteomic methods used gel-based methods of separating a protein mixture before analyzing the mixture with MS. Recently, multidimensional separation methods have been popular to separate peptides. Proteins are quantified through the incorporation of isotopic labels into cell culture, or through analyzing the mass spectra in the presence of a known reporter ion.
Isotopic labeling methods initially are achieved through incorporating chemical or metabolic labeling reagents into cell culture. One such method is the integration of isobaric tags for relative and absolute quantitation (iTRAQ). Researchers studying the Wnt/β-catenin signaling pathway were able to use iTRAQ to compare the proteomes of 2D to 3D monocultured SW480 cells [29]. Advanced tandem MS platforms coupled with multidimensional liquid chromatography enabled researchers to identify proteins by their diverse functions within biological samples. The use of iTRAQ allows for the relative abundance ratios of MS or MS/MS intensities of corresponding peptide pairs labeled with light-/heavy- isotope labels within the Wnt/B-catenin signaling pathway in this model.
The initial proteome analysis of 2D and spheroid cultures showed the differential expression of proteins within the Wnt/β-catenin signaling pathway. The researchers then treated the iTRAQ integrated 2D and 3D models with an inhibitor of the Wnt/β-catenin signaling pathway, known as XAV939. The comparison of 2D- and 3D-cultured SW480 cells showed a significantly statistical difference in protein abundance based on the fold change at a cut-off of 1.6. Although this study did not elucidate why XAV939 elicited this response within SW480 cells, the authors suggested a possible mechanism that may be related to the hypoxic conditions within 3D cell culture. They proposed that the hypoxic conditions contributed towards differential protein expression elicited by XAV939 [29]. This study did not utilize multiple TME components but provides a great example of a study that could alter its methodology in the early cell culture stages to include multiple TME components. The Wnt/B-catenin pathway is known to be modulated by stromal elements, and the addition of a stromal component could have allowed for a greater understanding of the biological processes that are affected the XAV939. With the stromal element present, the results would have also translated better to in vivo studies using the inhibitor.
More recently, a 2020 manuscript by Bauleth-Ramos used a triple co-culture with colorectal carcinoma cells, monocytes, and human intestinal fibroblasts to examine the molecular effect of nanoparticle treatments as a chemotherapeutics in vitro [29,55,56]. M2-like phenotypic macrophages are known to be important for tumor metastasis and poor prognosis resulting from monocytes. CAFs are also known to play a role in tumor progression and metastasis through the secretion of soluble factors and ECM proteins. Chemoimmunotherapy is often used as a promising approach to killing cancer. The goal of this study was to use nanoparticles to deliver chemoimmunotherapy to the triple cultured MCTS and determine the molecular processes that were altered by immunotherapy. Bauleth-Ramos seeded the carcinoma cell line HCT116, fibroblast HIF cells, and freshly isolated monocytes at a ratio of 4:1:4, respectively. The total number of cells initially seeded per spheroid was 5000, and 50 ng/mL of macrophage colony stimulating factor was supplemented to promote the differentiation of monocytes into macrophages within their culture environment.
Bauleth-Ramos found the triple co-culture of the MCTS was successful in adopting certain aspects characteristic of in vivo tumors, such as ECM developments, spatial organization, and the formation of a necrotic core. The model was phenotypically assessed using H&E staining, nanoparticle drug delivery effects were evaluated using fluorescent imaging, and flow cytometry worked to characterize the MCTS. Mono-, double- and triple-cultures revealed variations in tumor heterogeneity when compared. Drug resistance within the spheroids was found to be present and was illustrated by the stark difference in decreased penetration of nanoparticles from 2D cultures to MCTS. Despite significantly less uptake of the nanoparticles in the 3D triple co-culture model, proliferation was still decreased in the model compared to a negative control. In addition, macrophage polarization towards M1-like differentiation indicated anti-tumor behavior [57]. This study serves as an example of research that was able to elucidate relevant information about chemotherapeutic response using co-cultures. Nevertheless, if a similar study used MS-based proteomics, the conclusions made in this paper could be further validated, more robustly supported, and the biological mechanisms that drove their results could perhaps be further understood. The development of a triple co-culture model that assessed the immunologic response to a new chemotherapeutic was impressive; however, proteomics could have revealed the downstream effects that their nanoparticle delivery system or chemotherapy drug had on protein expression. This information could help to elucidate the molecular mechanism of how the chemotherapeutic elicited an anti-proliferation response.
The predominant method of co-culture is the use of MCTS with a single stromal component. However, even more complex models like triple co-cultures proceed to encapsulate more possibilities in TME recreation through the inclusion of multiple stroma. With a high mortality rate and few viable treatment options for pancreatic cancer patients, triple co-culture models are especially valuable in their ability to mimic the TME. A 2018 study conducted by Lazzari et al. provided one of the first uses of pancreatic MCTS in triple co-culture with fibroblasts and endothelial cells. This co-culture included pancreatic MCTSs derived from PANC-1 cells, human lung fibroblasts (MRC-5) and human umbilical vein endothelial cells. Novel aspects of the hetero-type, co-culture MCTS model appeared when tracking cell number, volume, and especially ATP content per spheroid. The two-fold difference in ATP in triple co-culture over mono-culture signified the stimulation of metabolic activity and contributed to resulting viability and increased survival rates upon treatment with doxorubicin [58].
3.2 |. Biomarker discovery
Biomarker discovery holds great clinical relevance, as it is extremely important that clinicians have reliable biomarkers that are consistent to allow for accurate diagnosis and staging. However, tumor heterogeneity of malignant tumors is quite often varied between individuals of the same cancer types. Genetic and epigenetic influences alter cellular phenotypes, which are reflected by subtle differences in molecular biology. Therefore, the identification of the cellular genotype and corresponding phenotype within cancer can help guide patient treatment and contribute to a better understanding of how oncogenic mutations alter the TME. MS is a global cellular analysis tool that can be useful for this purpose. In combination with data analysis methods, MS can be utilized as a beneficial tool for finding cellular identification patterns in large data sets, indicating it may be useful to identify potential biomarkers [59].
Proteomics studies intended to understand how biomolecular signaling drives malignant progression can serve as a platform for elucidating new biomarkers. In recent years, EVs have provided a resource to study communication that drives disease progression [60]. MS-based proteomics geared towards analyzing proteins held within these vesicles have gained prevalence for uncovering paracrine, autocrine, or juxtracrine signaling related to cancer formation and advancement. Cancer cells actively release EVs into neighboring tissue to facilitate communication. Exocytosis releases the vesicles from the cell, and it can be seen that EVs contain molecules that are released into cell junctions. Once isolated and analyzed, EVs have been found to carry proteins, lipids, metabolites, and RNAs, although the mechanisms through which these components enter exosomes are not fully understood. The use of cancer models using co-culturing could benefit from the understanding of mechanisms regarding EVs and further elucidate cellular mechanisms regarding communication in cancer progression. Co-cultures allow for natural interactions to be studied in a controlled environment [61]. EVs have been implicated in pathways involved in cell adhesion, migration, and protein transport. The mechanisms of interactions are not completely understood for every pathway, indicating the vast amount of information that is still left to be discovered. The scope of information that is unknown about communication in cancer makes it a useful research topic to initially explore in search of new potential biomarkers. LC/ESI-MS/MS is a prevalent analytical technique to study the contents of EVs. Proteomic analysis of EVs has shown significant alterations of proteins expressed under pathological and physiological conditions.
3.3 |. Elucidating the biochemistry of malignant progression
Studies have been performed in recent years that compare mono-cultured and co-cultured tumors to reveal differences in protein expression, appearance, cancer cell mobility, fundamental cell–cell interactions in the ECM [62,63]. Studies conducted by Jeong [62,63] and Kim [12] applied the co-culture of HT-29 colorectal MCTS with fibroblasts to further understand biological processes regarding malignant progression. Their studies emphasized the complex array of phenotypic benefits in mimicking the TME in co-culture over mono-culture. It is also important to note that each study had a different method of co-culture. Jeong et al. discovered that cell proliferation of HT-29 MCTS was increased when co-cultured with colorectal specific CAF cells (CDD-18Co) and cultured using a collagen scaffold. Colorectal MCTS exhibited a 1.5-fold increase in percentage change of the diameter after fibroblasts were added to the culture. Co-cultured MCTS exhibited a decrease in the expression of the proliferation marker Ki-67 of spheroids co-cultured with fibroblasts when compared to mono-cultured HT-29 MCTS, indicating that co-culture does alter the proliferation of spheroids [18].
Kim confirmed protein abundance changes in HT-29 MCTS co-cultured with CAF [54]. This study used non-MS-based proteomic methods to analyze co-cultured MCTS and study the implications of EMT in CRC. They used enzyme-linked immunosorbent assays to reveal differential expression of proteins involved in the TGF-β pathway. This pathway contributes to cell proliferation and differentiation. Upregulation of the growth factor proteins EGFR and CTGF was observed via western blot analysis, as well as the downregulation of β-catenin and E-cadherin. These changes validate that EMT was occurring within their model, and that processes known to be implicated in metastasis are also perturbed. Both Kim and Jeong’s models for EMT and tumor growth allowed a targeted proteomic analysis to reveal changes in protein expression levels to support their findings. However, it could have been improved if the global proteome was analyzed. Both studies relied on knowledge of currently known biological processes that are affected by cancer. Global proteomics analyses by MS could indicate what cellular pathways or signaling molecules are implicated in the perturbation of the spheroid growth with the addition of fibroblasts.
As emphasized previously, co-culture model systems better model the tumor environment to more closely resemble those in vivo. The spheroid model permits the idiosyncrasies of TME components to be amplified and more observable in co-culture. A recent example can be found in a study aimed at further understanding of the progression of ovarian cancer. Ovarian cancer progression relies on the interaction of cancer implanting in the mesothelium of the ovary, and the subsequent formation of invasive peritoneal implants [57]. Musrap co-cultured 2D ovarian cancer cells and peritoneal cells (LP-9), and collected the conditioned medium for LC-MS/MS analysis. The MS analysis detected 49 secreted proteins that varied in abundance from co-culture to mono-culture. Gene expression of Mucin 5A was elevated amongst three different ovarian cancer cell lines co-cultured with LP-9 mesothelial cells. These discoveries in co-culture gave a deeper understanding of mesothelium invasion, and identified pathways implicated in ovarian cancer metastasis (Figure 4). The study was limited by the use of 2D culture, as compared to if a 3D culture had been utilized. It would be interesting to repeat the experiments with co-cultured and mono-cultured spheroids.
FIGURE 4.
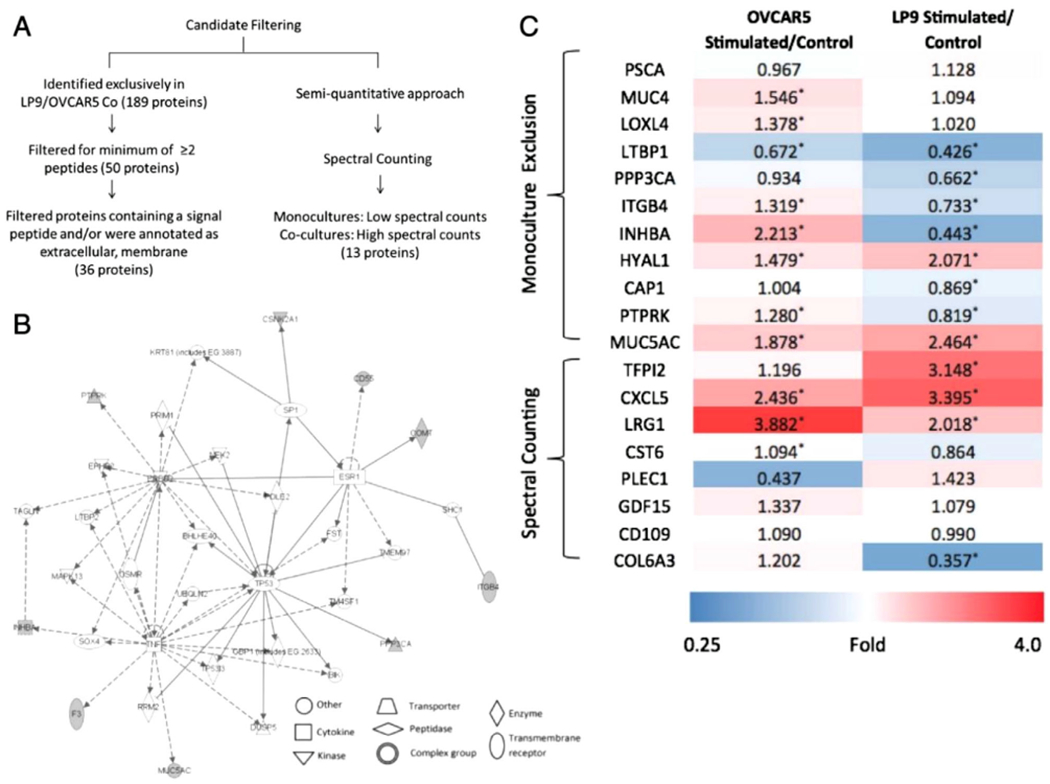
Global secretome analysis using LC-MS/MS, exploring the communication between ovarian cancer cell line, OVCAR-5, and mesothelial cell line, LP-9. (A) Filtering produced candidates of secreted proteins in the ECM. (B) IPA clustered candidate proteins in networks belonging to molecular transport, cancer, cell-to-cell signaling and interaction, and cell death and survival. Genes/proteins are depicted as nodes, with shaded nodes representing upregulated proteins, white nodes as genes/proteins incorporated by software to build networks. (C) Heatmap of mRNA expression of selected genes. Ratios represent fold changes in expression of stimulated cells over control cells. Red corresponds to increased gene expression, whereas blue illustrates reduced expression (*p ≤ 0.05, Student’s t-test). Reproduced from [29]
Microfluidic devices have been used to simulate TME. Although the devices vary and are often developed for specific experiments, they are useful in adding vascularization to models. As the MCTS models avascularized tumors, the addition of microfluidics to current MCTS models results in modeling a vascularized TME. Within the design of microfluidic devices, multiple channels will incorporate cancer cells while surrounding channels will possess media and components to deliver nutrients and oxygen as blood vessels would in the TME, with an example in Figure 5. In a study by Lee, a seven-channel microchannel plate was developed for investigation of chemoresistance and the EMT in co-cultures of pancreatic stellate cells and MCTS [29]. The microfluidic plate was designed with each channel alternating collagen matrix, media, and cells to simulate conditions of vascularization of tumors. A similar seven-channel microfluidic chip device was also separately employed by Jeong, modeling colorectal MCTS and CAF interactions [29,55,56]. In Lee’s work, immunofluorescence staining and a global MS proteome analysis of co-cultured MCTS revealed EMT-related markers. In each study, the multi-channel microfluidic co-culture allowed for recreation of ECM interactions between stromal and tumor components through a proximity model [57]. Additionally, in each study the use of MS-based proteomics could have enabled a better understanding of the processes occurring in each model. In Lee’s model, the EMT was studied; however, it would be valuable for a global proteomic analysis and subsequent pathway enrichment analysis to be performed to indicate the expression of all proteins involved in EMT-related pathways, rather than those that were targeted in this study.
FIGURE 5.
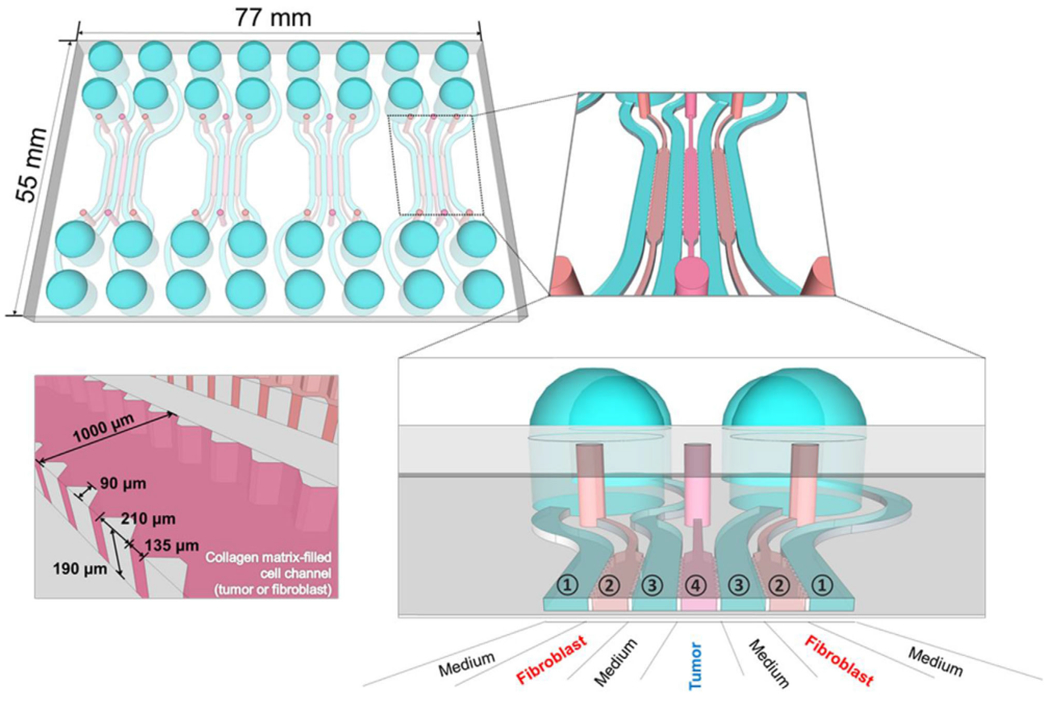
Microfluidic chip design structure and organization for co-culturing human colorectal cancer HT-29 cells with normal colorectal fibroblasts, CCD-18Co. The device had four units with even channels in each unit, with each channel being 1000μm in width. For mono-culture, the fibroblasts and collagen scaffold were fed into channel 2, and the cancer cells and collagen were fed into channel 4. For co-culture, a collagen suspension of HT-29 and CCD-18Co cells were loaded into channel 4. Media containing channel media separated each cell-containing channel and exposed the cells to nutrients while they were incubated in 5% CO2 and 37°C for 5 days to allow the spheroids in the device [57]
3.4 |. Cancer genome database contribution
In contrast to non-MS-based methods, MS is a severely underutilized tool to analyze 3D cancer models. MS data contains information regarding biochemical pathways that may be upregulated or down-regulated in co-cultured spheroids, yet the ability to interpret global proteomics data to understand what is occurring biologically is a major limiting factor shared by researchers. If global proteomic studies of co-cultures were validated and added to databases that are open access, the understanding of biological interactions in the TME could be greatly accelerated. Currently, a few databases exist including The Cancer Genome Atlas (TCGA), the Cancer Cell Line Encyclopedia (CCLE) and the Catalogue of Somatic Mutations in Cancer (COSMIC). TCGA is one of the first databases developed to characterize the macromolecular profiles of tumors and tumor cell lines. The project has inspired the foundation of other cancer databases, including COSMIC and CCLE. Databases are beneficial for capturing large amounts of data pertaining to cancer and can hold great potential. For example, a pan-cancer proteomic characterization of 532 cancers revealed gene signatures of oncogenic or metabolic pathways that can distinguish subtypes [58].
A Harvard group expanded the CCLE through quantitative profiling of 375 cell lines from diverse lineages to unearth previously unknown information. Within this massive effort, they used TMT10-plex labeling reagents and high-resolution tandem mass spectrometers. Their work resulted in 12,755 proteins identified and 4.7 million peptides. Their workflow included data mining, primary organization analysis, and proteogenomic analysis of MS1 and complexes. Data mining and PCA of the 4.7 million peptides indicated pathways across multiple cancers that were commonly differentially expressed. A large portion of the proteome was found to be correlated to epithelial and mesenchymal markers, and EpCam and Vimentin were identified as reliable epithelial and mesenchymal markers respectively across multiple tissue types. This study helped to validate the use of these proteins as biomarkers. Using proteogenomic analyses, they performed a PCA projection onto RNA data for cell lines with corresponding proteomic data. From this, they conclude that while RNA data can be correlated to proteomic data, the genomic material is not the primary component of variation of the steady state proteome. The vast extent of information uncovered in this study supports the idea that proteomics is a powerful tool capable of elucidating previously unknown information. Bioinformatics is a growing field, and it is possible that more information will be extracted from this data set in the future. This study did not culture the established cell lines in 3D, nor did they employ co-culture conditions. Yet this study serves as a beneficial example of how proteome wide analyses can yield insight into the TME.
Several studies have been performed that complement the CCLE or COSMIC. The research performed by Al-Juboori aimed to model endometrial cancer to further understand how cancerous and stromal tissue communication and cell to cell adhesion influence malignant progression. Their study co-cultured cancerous epithelial Ishikawa (IK) cells with non-cancerous human endometrium (HESC) into a MCTS model. To show the relevance of their model to protein expression in patient tumors, they co-cultured endometrial cells and epithelial cells isolated from extracted tumors. They phenotypically analyzed their model through fluorescent and brightfield imaging. They were able to observe that the RFP labeled IK cells formed layers around the GFP labeled HESC cells. The co-culture model was analyzed with LC-ESI-MS/MS. They identified a total of 1618 proteins in the co-cultured spheroids alone, and 500 proteins were found to be shared in the co-cultured, IK mono-cultured, and HESC mono-cultured spheroids. They performed an integrated pathway analysis to map common proteins within each co-cultured or monoculture population of the model and identify differentially expressed pathways (Figure 6A,B). Four pathways were found to be associated with proteins shared by both mono- and co-culture. These pathways were found to be implicated in significant protein-protein interactions, protein ubiquitination, regulation of eIF4 and p70S6k signaling, and mTOR signaling. The abundance of pathways that were found to be shared with both co-culture and monoculture provides biological relevance to their co-culture model and indicates its similarity to human normal and cancerous endometrium. Through using a combination MS and orthogonal methods, they were able to distinguish cell populations phenotypically and molecularly in their model and identify potential proteins that could serve as targets for future drug therapy (Figure 6C,D) [59].
FIGURE 6.
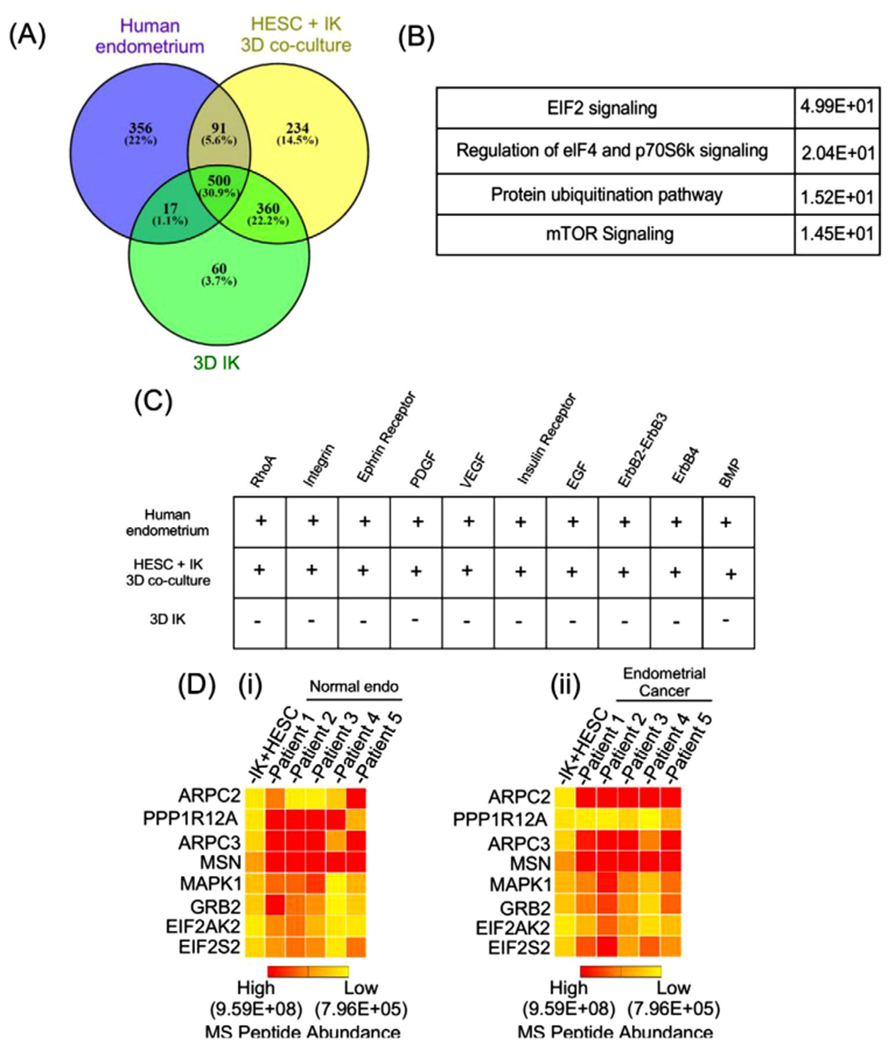
Proteome profile of stromal (human endometrium) and epithelial (IK) individual and co-cultured MCTS. (A) Venn diagram highlights the number of common and differently expressed proteins by LC-MS/MS in co-culture spheroids. (B, C) IPA of the proteome for commonly present signaling pathways in co-cultured MCTS. (D) Heat map. Reproduced from [29]
The addition of data from studies such as the one conducted by Al-Juboori to databases openly accessible could allow for an accelerated understanding of biological processes. Sharing data aids researchers in data storage, mining, and analysis. Shared databases provide bioanalysts with the opportunity to analyze data collected by skilled mass spectrometrists without publication restrictions and can serve as a future resource [64]. Al-Juboori’s work indicated several pathways that are potentially involved in malignant progression. Each pathway may perhaps serve as the basis of an individual research project. If the information contained in this study were to be added to COSMIC, the information could readily be accessible to those who are seeking proteomic data in cancer.
4 |. CONCLUSIONS
The intricacies of the TME stem from the interactions between the components. Stromal cells provide connective support for a 3D model of tumors and include endothelial cells and fibroblasts. During tumor development, normal stromal cells are constantly being recruited from the surrounding tissue. Once influenced by the cancerous cells, the stromal cells can express growth factors and mediators that aid in tumor growth, converting these components to be cancer related [62,63]. Much of the stroma is populated by fibroblast cells, which excrete signaling factors and proteins that can remodel the ECM. The ECM contains important enzymes, molecules, and signaling factors important for tumorigenesis and metastasis. Structures such as blood or lymphatic vessels provide nutrients and oxygen to the tumor.
While mono-clonal 3D cultures have been used as better models over 2D monolayers, they do not capture the full complexity of the TME. MCTS provide great potential for application with co-culture without the use of a scaffold and allow for direct cell-to-cell contact to occur encouraging the perturbation of the microenvironment similar to in vivo. MCTS as a model have the potential to be paired with microfluidics to model vascularized tumors. The use of MS for analyzing the proteomics of co-cultures is a severely underutilized technique. MS and MS imaging as analysis techniques for mono-cultures have proven to be extremely helpful in acquiring information about the proteome of the model in clinical applications, drug discovery, biomarker studies, cancer migration and invasion studies, and signaling [62,63]. Applying MS with co-culturing would open an avenue for greater high through-put, high-resolution molecular analysis.
Our goal in authoring this review article is to propose these valuable tumor models to the proteomics community and show their potential utility in exploring critical questions relating to cancer biology. We hope that as these models become more broadly available that their widespread proteomic analysis by MS will quickly follow suit. By pairing a more realistic tumor model with the exquisite multiplex chemical analysis possible through mass spectrometric methods, valuable discoveries that will ultimately benefit cancer patients would be enabled.
ACKNOWLEDGMENTS
ABH was supported by R01-GM110406. We thank Drs. Ralph Tobias and Emily Sekera for their careful reading of the manuscript and thoughtful suggestions.
Funding information
National Institute of General Medical Sciences, Grant/Award Number: R01-GM110406
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Hanahan D, & Weinberg RA (2011). Hallmarks of cancer: The next generation. Cell, 144, 646–649. [DOI] [PubMed] [Google Scholar]
- 2.Hawkins DS, Demers GW, & Galloway DA (1996). Cancer Research, 56, 892–895. [PubMed] [Google Scholar]
- 3.Soussi T, & Wiman KG (2015). TP53: An oncogene in disguise. Cell Death and Differentiation, 22, 1239–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xin X, Yang H, Zhang F, & Yang ST. (2019). 3D cell coculture tumor model: A promising approach for future cancer drug discovery. Process Biochemistry, 78, 148–149. [Google Scholar]
- 5.de Palma M, Biziato D, & Petrova TV (2017). Microenvironmental regulation of tumour angiogenesis. Nature Reviews Cancer, 17, 464–468. [DOI] [PubMed] [Google Scholar]
- 6.Aref AR, & Barbie D (2017). Cancer Drug Discovery and Development Ex Vivo Engineering of the Tumor Microenvironment, 1–8. [Google Scholar]
- 7.Koh B, Jeon H, Kim D, Kang D, & Kim KR (2019). Oncology Letters, 17, 2409–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoarau-Véchot J, Rafii A, Touboul C, & Pasquier J (2018). Halfway between 2D and animal models: are 3D cultures the ideal tool to study cancer-microenvironment interactions? International Journal of Molecular Sciences, 19, 181–194. 10.3390/ijms19010181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miki Y, Ono K, Hata S, Suzuki T, Kumamoto H, & Sasano H (2012). The advantages of co-culture over mono cell culture in simulating in vivo environment. Journal of Steroid Biochemistry and Molecular Biology, 131, 68–73. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, Kim EM, Byun H, kwan Chang H, Jeong K, Aman ZM, Choi YS, Park J, & Shin H (2018). Engineering spheroids potentiating cell-cell and cell-ECM interactions by self-assembly of stem cell microlayer. Biomaterials, 165, 105–106. [DOI] [PubMed] [Google Scholar]
- 11.Weiswald LB, Bellet D, & Dangles-Marie V (2015). Spherical cancer models in tumor biology. Neoplasia (United States), 17, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nath S, & Devi GR (2016). Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacology and Therapeutics, 163, 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fong ELS, Harrington DA, Farach-Carson MC, & Yu H (2016). Heralding a new paradigm in 3D tumor modeling. Biomaterials, 108, 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravi M, Paramesh V, Kaviya SR, Anuradha E, & Paul Solomon FD (2015). 3D cell culture systems: advantages and applications. Journal of Cellular Physiology, 230, 16–18. [DOI] [PubMed] [Google Scholar]
- 15.Fang Y, & Eglen RM (2017). SLAS Discovery, 22, 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cesarz Z, & Tamama K (2016). Spheroid culture of mesenchymal stem cells. Stem Cells International, 2016, 1–11. 10.1155/2016/9176357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Achilli TM, Meyer J, & Morgan JR (2012). Advances in the formation, use and understanding of multi-cellular spheroids. Expert Opinion on Biological Therapy, 12, 1347–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa EC, de Melo-Diogo D, Moreira AF, Carvalho MP, & Correia IJ (2018). Spheroids formation on non-adhesive surfaces by liquid overlay technique: considerations and practical approaches. Biotechnology Journal, 13, 1–10. 10.1002/biot.201700417 [DOI] [PubMed] [Google Scholar]
- 19.Kumar V, & Varghese S. (2019). Ex vivo tumor-on-a-chip platforms to study intercellular interactions within the tumor microenvironment. Advanced Healthcare Materials, 8, 1–9. 10.1002/adhm.201801198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patterson SD, & Aebersold RH (2003). Proteomics: The first decade and beyond. Nature Genetics, 33, 311–319. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, & Hummon AB (2015). Mass spectrometry imaging of therapeutics from animal models to three-dimensional cell cultures. Analytical Chemistry, 87, 9508–9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Flinders C, Mumenthaler SM, & Hummon AB (2018). MALDI mass spectrometry imaging for evaluation of therapeutics in colorectal tumor organoids. Journal of the American Society for Mass Spectrometry, 29, 516–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longuespée R, Casadonte R, Kriegsmann M, Pottier C, Picard de Muller G, Delvenne P, Kriegsmann J, & de Pauw E (2016). MALDI mass spectrometry imaging: A cutting-edge tool for fundamental and clinical histopathology. Proteomics Clinical Applications, 10, 701–704. [DOI] [PubMed] [Google Scholar]
- 24.Kim YE, Jeon HJ, Kim D, Lee SY, Kim KY, Hong J, Maeng PJ Kim K-R, & Kang D (2018). Quantitative proteomic analysis of 2D and 3D cultured colorectal cancer cells: Profiling of tankyrase inhibitor XAV939-induced proteome. Scientific Reports, 8, 13255–13261. 10.1038/s41598-018-31564-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauleth-Ramos T, Feijão T, Gonçalves A, Shahbazi MA, Liu Z, Barrias C, Oliveira MJ, Granja P, Santos HA, & Sarmento B (2020). Colorectal cancer triple co-culture spheroid model to assess the biocompatibility and anticancer properties of polymeric nanoparticles. Journal of Controlled Release, 323, 398–406. [DOI] [PubMed] [Google Scholar]
- 26.Wilkins MR, Sanchez J-C, Gooley AA, Appel RD, Humphery-Smith I, Hochstrasser DF, & Williams KL (1996). Progress with proteome projects: Why all proteins expressed by a genome should be identified and how to do it. Biotechnology & Genetic Engineering Reviews, 13, 19–25. [DOI] [PubMed] [Google Scholar]
- 27.Gygi SP, Corthals GL, Zhang Y, Rochon Y, & Aebersold R (2000). Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proceedings of the National Academy of Sciences of the United States of America, 97, 9390–9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JH, Kim SK, Khawar IA, Jeong SY, Chung S, & Kuh HJ (2018). Microfluidic co-culture of pancreatic tumor spheroids with stellate cells as a novel 3D model for investigation of stroma-mediated cell motility and drug resistance. Journal of Experimental and Clinical Cancer Research, 37, 1–8. 10.1186/s13046-017-0654-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Juboori AAA, Ghosh A, bin Jamaluddin MF, Kumar M, Sahoo SS, Syed SM, Nahar P, & Tanwar PS (2019). Proteomic analysis of stromal and epithelial cell communications in human endometrial cancer using a unique 3D co-culture Model. Proteomics, 19, 1–9. 10.1002/pmic.201800448 [DOI] [PubMed] [Google Scholar]
- 30.Frank SA, Iwasa Y, & Nowak MA (2003). Patterns of cell division and the risk of cancer number of surface cells regularly die and slough off. Genetics, 163, 1527–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu T, & Dai Y (2017). Tumor microenvironment and therapeutic response. Cancer Letters, 387, 61–65. [DOI] [PubMed] [Google Scholar]
- 32.Hinshaw DC, & Shevde LA (2019). The tumor microenvironment innately modulates cancer progression. Cancer Research, 79, 4557–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dongre A, & Weinberg RA (2019). New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nature Reviews Molecular Cell Biology, 20, 69–76. [DOI] [PubMed] [Google Scholar]
- 34.Devadas D, Moore TA, Walji N, & Young EWK. (2019).A microfluidic mammary gland coculture model using parallel 3D lumens for studying epithelial-endothelial migration in breast cancer. Biomicrofluidics, 13, 064122–064127>. 10.1063/1.5123912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SA, Lee EK, & Kuh HJ (2015). Co-culture of 3D tumor spheroids with fibroblasts as a model for epithelial-mesenchymal transition in vitro. Experimental Cell Research, 335, 187–194. [DOI] [PubMed] [Google Scholar]
- 36.Chae YK, Chang S, Ko T, Anker J, Agte S, Iams W, Choi WM, Lee K , & Cruz M (2018). Epithelial-mesenchymal transition (EMT) signature is inversely associated with T-cell infiltration in non-small cell lung cancer (NSCLC). Scientific Reports, 8, 2918–2926. 10.1038/s41598-018-21061-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bussard KM, Spaeth E, Mutkus LA, Stumpf KA, & Marini FC (2017). In Mesenchymal stromal cells as tumor stromal modulators. (pp. 253–273). Elsevier Inc. [Google Scholar]
- 38.Gieniec KA, Butler LM, Worthley DL, & Woods SL (2019). Cancer-associated fibroblasts-heroes or villains? British Journal of Cancer, 121, 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, Dekleva EN, Saunders T, Becerra CP, Tattersall IW, Westphalen CB, Kitajewski J, Fernandez-Barrena MG, Fernandez-Zapico ME, Iacobuzio-Donahue C, Olive KP, & Stanger BZ (2014). Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell, 25, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kahounová Z, Kurfürstová D, Bouchal J, Kharaishvili G, Navrátil J, Remšík J, Šimečková Š, Študent V, Kozubík A, & Souček K (2018). The fibroblast surface markers FAP, anti-fibroblast, and FSP are expressed by cells of epithelial origin and may be altered during epithelial-to-mesenchymal transition. Cytometry, Part A, 93, 941–950. [DOI] [PubMed] [Google Scholar]
- 41.Kalluri R (2016). The biology and function of fibroblasts in cancer. Nature Reviews. Cancer. 16, 582–598. 10.1038/nrc.2016.73 [DOI] [PubMed] [Google Scholar]
- 42.Bhome R, Mellone M, Emo K, Thomas GJ, Sayan AE, & Mirnezami AH (2018). In Methods in molecular biology (pp. 87–98). Humana Press Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo X, Oshima H, Kitmura T, Taketo MM, & Oshima M (2008). Stromal fibroblasts activated by tumor cells promote angiogenesis in mouse gastric cancer. Journal of Biological Chemistry, 283, 19864–19869. [DOI] [PubMed] [Google Scholar]
- 44.Folkman J (2007). Angiogenesis: An organizing principle for drug discovery? Nature Reviews Drug Discovery, 6, 273–281. [DOI] [PubMed] [Google Scholar]
- 45.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CHM, Jones DL, Visvader J, Weissman IL, & Wahl GM (2006). Cancer stem cells-perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Research, 66, 9339–9344. [DOI] [PubMed] [Google Scholar]
- 46.Paschos NK, Brown WE, Eswaramoorthy R, Hu JC, & Athanasiou KA (2015). Advances in tissue engineering through stem cell-based co-culture. Journal of Tissue Engineering and Regeneration Medicine, 9, 488–500. [DOI] [PubMed] [Google Scholar]
- 47.Visvader JE, & Lindeman GJ (2012). STEM, 10, 717–722. [DOI] [PubMed] [Google Scholar]
- 48.Yang Y (2015). Cancer immunotherapy: Harnessing the immune system to battle cancer. Journal of Clinical Investigation, 125, 3335–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pitt JM, Marabelle A, Eggermont A, Soria JC, Kroemer G, & Zitvogel L (2016). Targeting the tumor microenvironment: Removing obstruction to anticancer immune responses and immunotherapy. Annals of Oncology, 27, 1482–1486. [DOI] [PubMed] [Google Scholar]
- 50.Goers L, Freemont P, & Polizzi KM (2014). Co-culture systems and technologies: Taking synthetic biology to the next level. Journal of the Royal Society Interface, 11, 1–11. 10.1098/rsif.2014.0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Dijk M, Göransson SA, & Strömblad S (2013). Cell to extracellular matrix interactions and their reciprocal nature in cancer. Experimental Cell Research, 319, 1663–1670. [DOI] [PubMed] [Google Scholar]
- 52.Daley WP, Peters SB, & Larsen M (2008). Extracellular matrix dynamics in development and regenerative medicine. Journal of Cell Science, 121, 255–261. [DOI] [PubMed] [Google Scholar]
- 53.Krueger S, Kalinski T, Wolf H, Kellner U, & Roessner A (2005). Interactions between human colon carcinoma cells, fibroblasts and monocytic cells in coculture–regulation of cathepsin B expression and invasiveness. Cancer Letters, 223, 313–315. [DOI] [PubMed] [Google Scholar]
- 54.Jeong SY, Lee JH, Shin Y, Chung S, & Kuh HJ (2016). Co-culture of tumor spheroids and fibroblasts in a collagen matrix-incorporated microfluidic chip mimics reciprocal activation in solid tumor microenvironment. PLoS One, 11, 1–11. 10.1371/journal.pone.0159013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim S, Kim S, Kim J, Kim B, Kim SI, Kim MA, Kwon S, & Song YS (2018). Evaluating tumor evolution via genomic profiling of individual tumor spheroids in a malignant ascites. Scientific Reports, 8, 2–8. 10.1038/s41598-018-31097-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kriegsmann M, Longuespée R, Wandernoth P, Mohanu C, Lisenko K, Weichert W, Warth A, Dienemann H, de Pauw E, Katzenberger T, Aust D, Baretton G, Kriegsmann J, & Casadonte R (2017).Typing of colon and lung adenocarcinoma by high throughput imaging mass spectrometry. Biochimica et Biophysica Acta - Proteins and Proteomics, 1865, 862–864. [DOI] [PubMed] [Google Scholar]
- 57.Wit M, Fijneman RJA, Verheul HMW, Meijer GA, & Jimenez CR (2013). Proteomics in colorectal cancer translational research: Biomarker discovery for clinical applications. Clinical Biochemistry, 46, 468–475. [DOI] [PubMed] [Google Scholar]
- 58.Chen F, Chandrashekar DS, Varambally S, & Creighton CJ (2019). Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nature Communications, 10, 3–7. 10.1038/s41467-019-13528-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schatoff EM, Leach BI, & Dow LE (2017). WNT signaling and colorectal cancer. Current Colorectal Cancer Reports, 13, 105–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bandu R, Oh JW, & Kim KP (2019). Mass spectrometry-based proteome profiling of extracellular vesicles and their roles in cancer biology. Experimental and Molecular Medicine, 51, 2–7. 10.1038/s12276-019-0218-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimasaki T, Yamamoto S, & Arisawa T (2018). Exosome research and co-culture study. Biological & Pharmaceutical Bulletin, 41, 1315–1319. [DOI] [PubMed] [Google Scholar]
- 62.Zink KE, Dean M, Burdette JE, & Sanchez LM (2018). Imaging mass spectrometry reveals crosstalk between the fallopian tube and the ovary that drives primary metastasis of ovarian cancer. ACS Central Science, 4, 1361–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zink KE, Dean M, Burdette JE, & Sanchez LM (2019). Capturing small molecule communication between tissues and cells using imaging mass spectrometry. Journal of Visualized Experiments, 2019, 2–7. 10.3791/59490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Creighton CJ, & Duncan DL (2018). Curr Protoc Mol Bio, 121, 19.14.1–19.14.13. 10.1002/cpmb.49 [DOI] [PMC free article] [PubMed] [Google Scholar]


