ABSTRACT
Members of the Corynebacterineae suborder of bacteria, including major pathogens such as Mycobacterium tuberculosis, grow via the insertion of new cell wall peptidoglycan (PG) material at their poles. This mode of elongation differs from that used by Escherichia coli and other more well-studied model organisms that grow by inserting new PG at dispersed sites along their cell body. Dispersed cell elongation is known to strictly require the SEDS-type PG synthase called RodA, whereas the other major class of PG synthases called class A penicillin-binding proteins (aPBPs) are not required for this mode of growth. Instead, they are thought to be important for maintaining the integrity of the PG matrix in organisms growing by dispersed elongation. In contrast, based on prior genetic studies in M. tuberculosis and related members of the Corynebacterineae suborder, the aPBPs are widely believed to be essential for polar growth, with RodA being dispensable. However, polar growth has not been directly assessed in mycobacterial or corynebacterial mutants lacking aPBP-type PG synthases. We therefore investigated the relative roles of aPBPs and RodA in polar growth using Corynebacterium glutamicum as a model member of Corynebacterineae. Notably, we discovered that the aPBPs are dispensable for polar growth and that this growth mode can be mediated by either an aPBP-type or a SEDS-type enzyme functioning as the sole elongation PG synthase. Thus, our results reveal that the mechanism of polar elongation is fundamentally flexible and, unlike dispersed elongation, can be effectively mediated in C. glutamicum by either a SEDS-bPBP or an aPBP-type synthase.
KEYWORDS: apical growth, cell wall, morphogenesis, peptidoglycan
INTRODUCTION
Virtually all bacterial cells are surrounded by a cell wall made of peptidoglycan (PG). This heteropolymer consists of glycan chains with a disaccharide repeat and a short peptide chain attached to one of the sugars (1). The peptides are used to generate amide cross-links between glycans, which ultimately allow the formation of an interconnected meshwork that surrounds the cell and protects its membrane from osmotic rupture (1). Synthesis of the PG matrix requires two enzymatic activities: glycosyltransferases (GTases) that polymerize the glycan strands and transpeptidases (TPases) that cross-link the peptide side chains (1). The PG synthases possessing these activities come in two forms. Class A penicillin-binding proteins (aPBPs) are the most well-studied synthase type (1). They are bifunctional and possess both GTase and TPase activities in a single polypeptide (2–5). The second type of synthase was discovered more recently and is formed through an interaction between a SEDS (shape, elongation, division, and sporulation) family GTase and a monofunctional class B PBP (bPBP) with TPase activity (6–10). Despite the importance of these enzymes for cell growth and morphogenesis and their status as key antibiotic targets, we are only just beginning to understand how they work together to build and expand the cell-sized PG matrix called the sacculus that defines cell shape.
Bacciliform or rod shape is a common bacterial morphology. It is generated via one of two distinct modes of cell wall growth: dispersed synthesis or apical/polar elongation (11) (Fig. 1). Dispersed PG elongation has been more widely studied because it is the growth mode employed by the major Gram-negative and Gram-positive model systems, Escherichia coli and Bacillus subtilis. This growth mode is carried out by a highly conserved protein complex called the Rod system (elongasome) (1). The essential PG synthase of this system is a SEDS-bPBP complex formed between RodA and PBP2 (9). PG synthesis by this enzyme complex is spatially controlled via its connection to filaments of the actin-like MreB protein, which are thought to promote the insertion of new PG glycans at many sites around the cell cylinder in an orientation orthogonal to the long cell axis (12–14). The Rod complex also includes the membrane protein components RodZ, MreC, and MreD (15–19). Although the function of these components is still being investigated, evidence is accumulating that they are involved in modulating MreB polymerization in the case of RodZ (20) or the activation of the RodA-PBP2 synthase in the case of MreC (9). Inactivation of any of the Rod system components, including the RodA-PBP2 synthase, results in the loss of rod shape and is often lethal to organisms that grow via this dispersed mode of elongation (21).
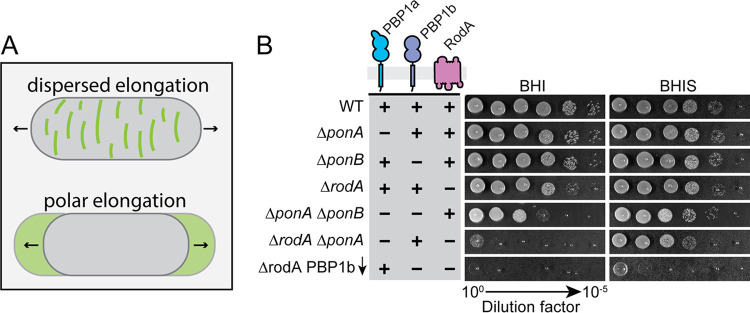
FIG 1.
Growth phenotype of mutants inactivated for one or more elongation PG synthase. (A) Schematic highlighting the two different modes of cell elongation used by rod-shaped cells. Green color depicts areas of new cell wall insertion. (B) Overnight cultures of MB001 (WT) and its indicated derivatives HL18 (ΔponA), JS20 (ΔponB), HL31 (ΔrodA), JS36 (ΔponA ΔponB), JS38 (ΔrodA ΔponA), and JS21(attB1::pJWS117) (ΔrodA ΔponB [Psod::riboE1-mScar-ponB]) grown in BHIS medium at 30°C were normalized to an OD600 of 0.5 and serially diluted. An aliquot of each dilution (5 μl) was then spotted onto BHI or BHIS agar medium. Plates were incubated at 30°C for 24 h and photographed. The expression construct pJWS117 encodes an mScar-PBP1b fusion with the theophylline-inducible riboE1 riboswitch (50) controlling its translation. Inducer was not added to the medium in order to deplete JS21(attB1::pJWS117) cells of PBP1b.
The aPBPs are also thought to be involved in dispersed cell elongation. However, simultaneous inactivation of the two major aPBPs in E. coli and other Gram-negative bacteria results in rapid cell lysis instead of the gradual loss of shape observed when Rod system function is disrupted (22). Moreover, the aPBPs are entirely dispensable for cell growth in B. subtilis, and their activity has been associated with the production of PG material that is less oriented in structure than that produced by the Rod system (23, 24). Additionally, the localization dynamics of the aPBPs have been observed to be distinct from the proteins of the Rod system (7). Thus, the emerging picture is that the aPBPs are likely to be working outside the Rod complex and may participate in cell elongation by filling voids in the oriented PG structure produced by the MreB-directed RodA-PBP2 synthase.
Growth via the insertion of new PG material at the cell poles is an alternative mechanism by which rod shape can be generated (11). This growth mode is used by some alphaproteobacteria such as Agrobacterium tumefaciens (25). It is also the major growth mode of the Actinobacteria phylum, including members of the Corynebacterineae suborder such as Mycobacterium tuberculosis (Mtb), Mycobacterium smegmatis (Msmeg), and Corynebacterium glutamicum (Cglu) (26, 27). Mtb and its relatives lack orthologs of all Rod system components except for the RodA-PBP2 synthase. Instead of MreB, a cytoskeletal-like structure formed by the DivIVA (Wag31) protein at the cell poles is thought to organize the polar growth machinery (27–30). Although both classes of cell wall synthases localize to the cell poles (30–33), their relative roles in promoting apical growth remain unclear. In contrast to organisms that grow via dispersed elongation, RodA and PBP2 are not essential for cell elongation in the mycobacteria or Cglu (30, 33, 34). The essentiality of the aPBPs also varies among these organisms. In Msmeg, one of its two major aPBPs (PonA1/PBP1) is essential for growth, and its depletion leads to lysis and severe morphological defects (31, 35), whereas in Mtb, the major aPBPs are individually dispensable but form a synthetic lethal pair (36). Surprisingly, Cglu mutants lacking their major aPBPs were found to be viable, but the cells displayed a spherical shape (33). Based on these genetic results, polar growth in Corynebacterineae is generally assumed to require aPBP function. However, the process of polar growth has not been directly assessed in mutants of these organisms lacking aPBP activity. We therefore decided to reinvestigate the relative roles of the different types of PG synthases in morphogenesis and polar growth in Corynebacterineae using Cglu as a model system.
Cglu harbors three enzymes with PG polymerase activity that localize to the cell poles and are likely to participate in cell elongation: two aPBPs, PBP1a and PBP1b, and the SEDS protein RodA (30, 33). Surprisingly, the construction of individual and multiple deletion mutants inactivating these factors revealed that polar growth can be mediated by either type of synthase in the absence of the other. Furthermore, subcellular localization studies indicate that the aPBPs and RodA have different spatial distributions at the cell poles, suggesting that despite their individual sufficiency for polar growth, they may play distinct roles in the process. Overall, our results reveal that the mechanism of polar elongation is fundamentally flexible and, unlike dispersed cell elongation, can be effectively mediated in Cglu by either a SEDS-bPBP or an aPBP-type synthase.
RESULTS
Phenotypic analysis of Cglu mutants lacking a single PG synthase.
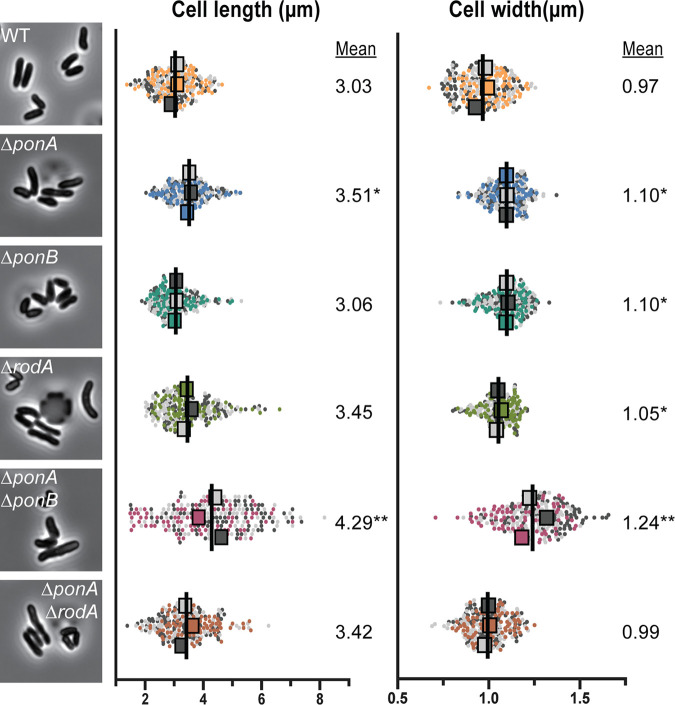
To investigate the relative contributions of the different PG synthases in morphogenesis and polar growth, the corresponding genes were first deleted individually. Consistent with previous findings (30, 33), mutants lacking any single synthase were viable (Fig. 1). Deletion of either ponA or ponB, encoding the aPBP-type enzyme PBP1a or PBP1b, respectively, did not result in a substantial growth or shape defect (Fig. 1 and 2). The only observable change was that ΔponA cells were slightly longer than wild-type cells (Fig. 2). A previous phenotypic profiling analysis indicated that mutants inactivated for PBP1a or PBP1b are hypersensitive to the cell wall-targeting antibiotics ampicillin or meropenem, respectively (37). These sensitivities displayed by the ΔponA or ΔponB mutant strains were complemented by the production of the missing synthase fused to the fluorescent protein mScarlet (mScar) (38), indicating that the phenotypes of the deletion mutants are not due to adverse effects on the expression of nearby genes (see Fig. S1A and B in the supplemental material).
FIG 2.
Morphological analysis of mutants lacking elongation PG synthases. Overnight cultures of the indicated strains from Fig. 1 grown in BHIS medium at 30°C were diluted 1:1,000. The cultures were then grown to an OD600 of 0.1 in BHIS medium at 30°C. Cells from the cultures were subsequently loaded into CellASIC devices and imaged by phase-contrast microscopy. Representative micrographs of each strain are shown adjacent to SuperPlots (51) of the corresponding morphological measurements. Cell length and width were measured for three replicate populations of 80 cells for a total of 240 cells for each strain using Oufti (48). The mean from each replicate population (represented by the large squares whose color corresponds to the replicate population from which it was calculated) was used for statistical tests. Analysis of variance (ANOVA) tests comparing replicate means to WT means were performed. *, P < 0.05; **, P < 0.0001.
Complementation of PG synthase mutant phenotypes. (A to C) Overnight cultures of MB001 (WT) and the indicated derivatives HL18 (ΔponA), JS20 (ΔponB), and HL31 (ΔrodA) grown in BHIS medium at 30°C were normalized to an OD600 of 0.5, serially diluted 10-fold, and spotted (5 μl) onto BHI or BHIS agar medium supplemented with theophylline (0.3 mM), ampicillin (0.2 μg/ml), or meropenem (0.03 μg/ml) where indicated. Plates were incubated for 24 h at 30°C and photographed. Plasmids used were pJWS19 (Psod::riboE1-mScar-ponA Kanr) and pJWS116 (Psod::riboE1-mScar-ponA Aprar] (A), pJWS94 (Psod::riboE1-mScar-ponB Kanr) and pJWS117 (Psod::riboE1-mScar-ponB Aprar) (B), and pJWS33 (Psod::riboE1-rodA-mScar Kanr) (C). Download FIG S1, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Compared to the strains lacking one of the two major aPBPs, the rodA deletion mutant displayed more pronounced growth and shape defects. As observed previously, ΔrodA cells form smaller colonies than the wild-type control on brain heart infusion (BHI) medium at 30°C and fail to grow at 37°C (30) (Fig. 1 and S1C). However, in contrast to the prior report, which found Cglu cells lacking RodA to be shorter and rounder than normal (30), we observed a broad distribution of cell lengths for the ΔrodA mutant, with subpopulations of cells that were shorter and longer than the wild type. Overall, the average cell length of the mutant increased relative to that of the wild type (Fig. 2), and this increase in length was associated with the observation of unresolved septa in ΔrodA cells, indicating a defect in cell separation (see Fig. S2). A similar phenotype was previously observed for mutants lacking PBP2b (33), consistent with it being the likely bPBP partner for the RodA polymerase. Why these phenotypes are different from the previous analysis of rodA mutants is not clear, but it might be due to differences in the growth phase at which the cells were visualized. The shorter cells in the prior report (30) may have been imaged at a later phase of growth, potentially allowing resolution of the incomplete septa and the generation of a shorter-than-average population of cells. Nevertheless, because the phenotype of our rodA deletion mutant is similar to that caused by PBP2b inactivation (33) and this phenotype can be complemented by the ectopic production of a Rod-mScar fusion (Fig. S2), we conclude that cells defective for the RodA-PBP2b synthase have a cell separation defect. This finding suggests that the proper assembly of the future cell elongation machinery at the developing daughter cell pole may help complete the process of cell division (see Discussion).
Cell division phenotype of ΔrodA cells and complementation by mScar-RodA. (A) Representative phase contrast micrographs of MB001 (WT) and its HL31 (ΔrodA) derivative. Overnight cultures grown in BHIS medium at 30°C were diluted 1:1,000 in BHI medium and grown at 30°C. When the OD600 reached 0.2 to 0.3, cells were diluted 10-fold and loaded into a CellASIC microfluidic device and imaged by phase-contrast microscopy. Where indicated, RodA-mScar production was induced from integrated pJWS33 (Psod::riboE1-rodA-mScar Kanr) with theophylline (0.3 μM). The yellow arrowhead indicates longer cells within the population, while cyan arrowheads indicate unusually small cells that originated from recently v-snapped long cells. (B) Representative fluorescence micrographs of the strains from panel A grown as described above but pulsed labeled with TADA for 5 min prior to imaging. (C) Histograms showing cell length distributions of the ΔrodA mutant with or without RodA-mScar production. Download FIG S2, TIF file, 1.0 MB (985.8KB, tif) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Polar elongation does not require aPBP activity.
We next investigated the phenotypes of mutants lacking multiple PG synthases. Consistent with prior results (35), we were able to construct a double ΔponA ΔponB mutant of Cglu devoid of all aPBPs. Although the mutant was viable and plated with near normal efficiency on BHI medium, the resulting colonies were much smaller than those of the wild type or the mutants lacking a single aPBP (Fig. 1). Notably, the addition of sorbitol to the BHI medium (denoted as BHIS) significantly improved the growth of the double ΔponA ΔponB mutant, likely by osmotically protecting the cells (Fig. 1). Strikingly, when we imaged mutant cells growing under these conditions (Fig. 2) or even in plain BHI medium (see Fig. S3), their morphology was very different from that observed in the prior analysis of a double ΔponA ΔponB mutant, in which a single image of a small number of spherical cells was shown (35). The type of growth medium used for the prior experiment was not clearly indicated nor was a population-level analysis of cell morphology performed, but the observed spherical shape of the mutant has contributed to the commonly held belief that aPBP activity is critical for polar elongation. In contrast, our morphological analysis of ΔponA ΔponB mutant cells growing in BHIS medium revealed that they are quite capable of growing with a long axis, albeit with a much broader length distribution and an increased average width relative to that of wild-type cells (Fig. 2).
Morphology of ΔponA ΔponB cells grown in BHI medium without sorbitol. An overnight culture of JS36 (ΔponA ΔponB) grown in BHIS medium at 30°C was diluted 1:1,000 in BHI medium and grown at 30°C. When the OD600 reached 0.1, cells were loaded on BHI agar pads and imaged by phase-contrast microscopy. Download FIG S3, TIF file, 0.6 MB (659.8KB, tif) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To assess the ability of the ΔponA ΔponB mutant to elongate from the cell pole, we performed a pulse-chase labeling experiment with the fluorescent d-amino acid (FDAA) TADA (39), which labels the PG layer (Fig. 3). Cells were first grown in the presence of TADA to label their cell walls. The label was then removed from the medium, and the cells were imaged over time by both phase-contrast and fluorescence microscopy. During this chase period, newly incorporated PG material can be monitored by the appearance of nonfluorescent areas of cell mass. As expected, for cells of the wild-type strain and mutants lacking a single PG polymerase, regions of new wall growth appeared at the cell poles (Fig. 3). The aPBP-less mutant similarly displayed the ability to grow through the addition of new PG material at the cell poles (Fig. 3). Notably, TADA labeling also revealed that this mutant had an even more severe cell separation defect than the ΔrodA strain, with ∼20% of cells in the ΔponA ΔponB population displaying multiple septa relative to ∼5% for cells lacking RodA (Fig. 4). Based on this analysis, we conclude that aPBPs are not required for polar growth in Cglu and that the SEDS-bPBP synthase RodA-PBP2b is likely sufficient to promote this mode of PG elongation. Additionally, the cell separation phenotype of the ΔponA ΔponB mutant underscores the connection between cell elongation PG synthases and the final stages of cell division in this bacterium.
FIG 3.

RodA and PBP1b promote cell growth by inserting nascent PG material at the poles. Overnight cultures of the indicated strains from Fig. 1 grown in BHIS medium at 30°C were diluted 1:1,000 in BHIS and grown for 3 h at 30°C. The cells were loaded into a CellASIC microfluidic device and grown for 30 min in BHIS medium at 30°C. Following this equilibration period, the cells were imaged every 5 min using phase-contrast and fluorescence optics. Cells were pulse labeled with the fluorescent d-amino acid TADA (39) for 3 min at the 6-min mark. The label was then progressively washed away by the flow of fresh medium lacking label. Every 6th frame in the time-lapse series is shown. The length of unlabeled cell wall was measured after the TADA pulse (t = 20 min) and at the end of the time lapse (t = 120 min) using Oufti (N > 150 cells); the unlabeled portion of each cell was defined as that with a TADA signal <20% of the maximum TADA signal for that cell. The mean and standard error of the rate of polar elongation were calculated by taking the mean difference in the length of unlabeled cell wall at the initial and final time points divided by the time elapsed.
FIG 4.
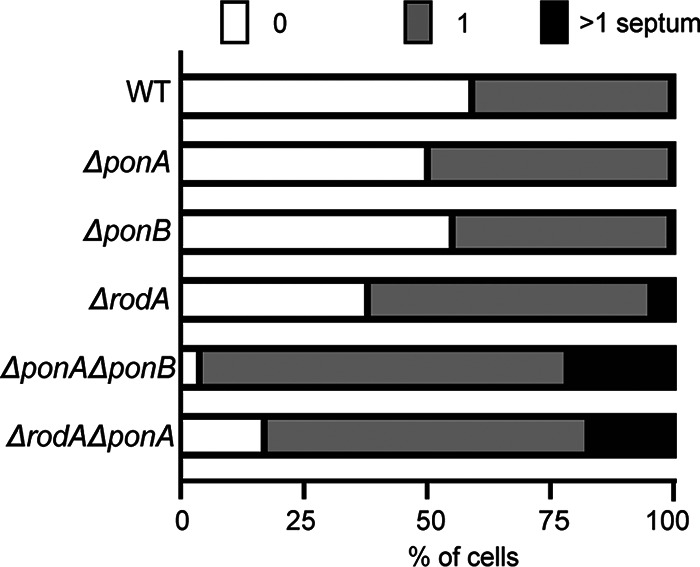
Some mutants lacking PG synthases have a cell separation defect. Using images from the 20-min time point of the time-lapse experiment in Fig. 3, the number of TADA-labeled septa in cells (n > 250) of each strain were quantified using the Cell Counter plugin in FIJI. The percentages of cells displaying no septa, one septum, or multiple septa are shown for each strain.
PBP1b can promote polar growth in the absence of other elongation synthases.
In addition to the ΔponA ΔponB mutant, we also sought to construct and characterize double mutants lacking RodA along and one of the two aPBPs. We were unable to construct either mutant on BHI medium but were successful in generating the ΔponA ΔrodA mutant on BHIS medium. It was only possible to construct the ΔponB ΔrodA mutant when an mScar-PBP1b fusion was ectopically expressed from a theophylline-inducible construct. Viability analysis confirmed that both the ΔponA ΔrodA mutant and the ΔponB ΔrodA strain depleted of PBP1b had a severe plating defect on BHI medium (Fig. 1). This plating defect was corrected on BHIS medium for the strain deleted for ponA and rodA, but even under these conditions, the mutant displayed a slow-growth phenotype relative to that of the wild type based on colony size (Fig. 1). The morphology of the ΔponA ΔrodA mutant in BHIS medium was very similar to that of cells lacking RodA alone (Fig. 2). The cells were mildly elongated relative to wild-type cells and retained the ability to elongate from the cell poles (Fig. 3). They also displayed a cell separation defect that was increased in severity compared to that of the single rodA deletion mutant (Fig. 4). Because the ΔponB ΔrodA mutant stopped growing almost immediately following PBP1b depletion, even when cultured in BHIS medium, we were unable to characterize its morphology in a manner similar to that for the other mutants in the collection.
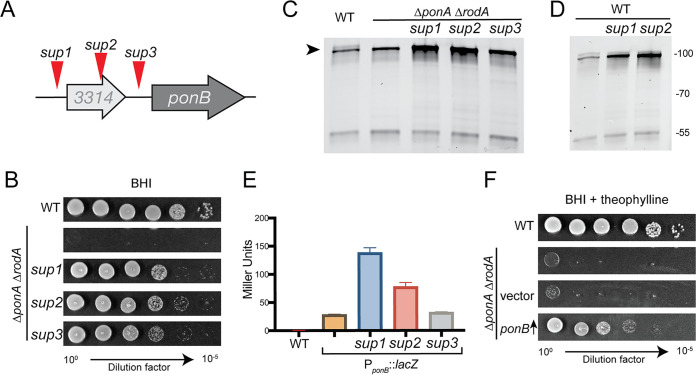
To investigate why the ΔponA ΔrodA mutant required sorbitol supplementation for growth, it was plated on plain BHI medium to isolate spontaneous suppressors that grew in the absence of sorbitol. Several survivors were purified, confirmed to have gained the ability to grow on BHI medium, and subjected to whole-genome sequencing to map the mutations causing the suppressor phenotype. Several of the isolated suppressors had one of three possible mutations near ponB (Fig. 5A and B). One was upstream (sup1) and one was within (sup2) the cgp_3314 gene, which is found just upstream of ponB, and the third mutation (sup3) was located in the intergenic region between cgp_3314 and ponB.
FIG 5.
Increased PBP1b production suppresses the growth defect of ΔrodA ΔponA cells. (A) Diagram of the ponB locus showing the approximate location of the suppressor mutations. The mutations mapped to nucleotide positions 2,959,545, 2,959,217, or 2,959,117 for sup1, sup2, or sup3, respectively, and all were G-to-A transitions. The sup1 strain had one additional mutation in tetA, the sup2 strain had no additional mutations, and the sup3 strain had additional mutations in fkpA, cgtR5, upstream of pitA and gdh, and downstream of phoD. (B) Overnight cultures of the ΔrodA ΔponA strain (JS38) and its sup1, sup2, and sup3 derivatives were grown in BHIS medium at 30°C. Culture densities were normalized to an OD600 of 0.5. The normalized cultures were then serially diluted and plated as described in the legend for Fig. 1. Cells were grown on BHI medium at 30°C for 24 h before the plates were photographed. Cells of the strains from panel B (C) or MB001 (WT) and its engineered derivatives containing the sup1 or sup2 mutation in the ponB locus (D) were labeled with Bocillin-FL. Membrane extracts were then prepared from the labeled cells, and 5 μg of total protein for each was subjected to SDS-PAGE. Bocillin-FL-labeled proteins were then detected using a Typhoon 9500 imager with excitation at 488 nm and emission at 530 nm. (E) Overnight cultures of MB001 (WT) with or without a plasmid containing the indicated wild-type or mutant ponB locus fused to a promoterless lacZ reporter were grown in BHIS medium at 30°C. They were diluted 1:200 and grown to an OD600 of 0.3 under the same conditions. Cell pellets were then collected and frozen. Thawed cells were then resuspended in Z buffer and permeabilized with toluene and lysozyme. o-Nitrophenyl-β-d-galactopyranoside (ONPG) was added, CaCO3 was used to stop the reaction, and the absorbance of the reaction measured at 420 nm. Beta-galactosidase activity was determined using the equation activity = [OD420 − 1.75 (OD550)]/[time (min) × 1 ml vol × OD600 × 1,000]. (F) MB001 (WT) and its ΔrodA ΔponA derivative with no plasmid, an empty vector, or a vector expressing mScar-PBP1b (pJWS117) were grown and plated as for panel B. The cells were grown on BHI medium supplemented with theophylline to induce PBP1b production in the pJWS117-containing strain.
The location of the suppressor mutations upstream of ponB suggested that they may be affecting its expression. Accordingly, when PBP1b levels were assessed in the ΔponA ΔrodA mutant and the suppressors using the fluorescent beta-lactam Bocillin-FL that covalently modifies the TP active site, an increase in the abundance of PBP1b was detected in the suppressor strains relative to that in the wild type and the ΔponA ΔrodA parent strain (Fig. 5C andS4). Two of the suppressor mutations were reconstructed in an otherwise wild-type background, and both also led to an increase in PBP1b levels, indicating that the observed change does not require PBP1a or RodA to be inactivated (Fig. 5D). To determine whether the suppressor mutations were affecting ponB transcription, we constructed a plasmid with the region upstream of ponB, including cgp_3314 and its upstream region, in front of a promoter-less lacZ reporter gene. Following introduction of this plasmid into the wild-type strain with or without the sup1 or sup2 changes at the chromosomal ponB locus, no change in LacZ activity was observed (not shown). However, addition of the sup1 or sup2 but not sup3 changes to the plasmid resulted in an increase in LacZ activity (Fig. 5E). Thus, the suppressors only appear to be effective when they are in cis, with sup1 and sup2 causing enhanced ponB transcription. The remaining suppressor, sup3, potentially affects ponB translation as opposed to transcription given its limited effect on lacZ expression in the reporter construct.
Bocillin-labeling controls for determining PBP identity. Cells of MB001 (WT) and its derivatives HL18 (ΔponA), JS20 (ΔponB), JS12 (Δpbp2a), and HL37 (Δpbp2b) labeled with Bocillin-FL. Membrane extracts were then prepared from the labeled cells, and 5 μg of total protein for each was subjected to SDS-PAGE. Bocillin-FL-labeled proteins were then detected using a Typhoon 9500 imager with excitation at 488 nm and emission at 530 nm. (A) Gel was run at 110 V for 1.5 h. (B) Gel was run at 110 V for 3 h to help resolve the multiple bands at running at ∼100 kDa. Download FIG S4, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Although the mechanism by which the suppressors are affecting ponB expression remains to be determined, the genetic analysis suggested that the main reason that the ΔponA ΔrodA mutant cannot grow on BHI medium is not due to the loss of PBP1a or RodA per se but rather results from a general deficit in PG synthase activity. Accordingly, the overproduction of mScar-PBP1b alone was sufficient to restore growth of the ΔponA ΔrodA mutant on BHI medium (Fig. 5F) and promote a morphology that closely matched that of the original suppressor mutants and wild-type cells (see Fig. S5). We therefore conclude that just as RodA-PBP2b appears to be sufficient to promote polar growth in the absence of the aPBPs, the aPBP-type enzyme PBP1b is also capable of promoting this mode of growth in the absence of the other elongation PG synthases. Thus, unlike dispersed cell elongation, polar elongation can be mediated by either a SEDS-bPBP synthase or an aPBP.
Morphology of cells with PBP1b as their only elongation PG synthase. Overnight cultures of MB001 (WT) and the indicated derivatives of JS38 (ΔponA ΔrodA) grown in BHIS medium at 30°C were diluted 1:1,000. The cultures were then grown to an OD600 of ∼0.05 before being loaded into CellASIC devices for imaging by phase-contrast microscopy. Cell length and width were measured for a population of cells of each strain (n > 240) using Oufti. Representative micrographs of each strain are shown adjacent to plots of the corresponding morphological measurements. Each dot in the plots represents measurements from a single cell. The lines indicate the mean measurement plus or minus one standard deviation. The medium was supplemented with theophylline (0.6 mM) to induce mScar-PBP1b production in the pJWS117-containing strain. Download FIG S5, TIF file, 2.5 MB (2.5MB, tif) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PG synthase inactivation is not accompanied by increased transcription of genes encoding another synthase.
In B. subtilis, viability of the strain lacking all aPBPs depends on a stress response that increases the expression of rodA, presumably because excess RodA is required to compensate for the loss of the aPBP-type PG polymerases (6). This result along with our finding that suppressors of the ΔponA ΔrodA mutant enhance ponB expression prompted us to investigate whether the viability of any of the deletion mutants we constructed depended on a transcriptional response that altered the expression of genes encoding the remaining PG synthases. We therefore performed transcriptome analysis (RNA-seq) on each of the deletion mutants and assessed whether the expression level for any of the genes encoding aPBP, bPBP, or SEDS proteins was altered by a deletion or pair of deletions. The only significant changes that were observed were in the expression of the deleted gene itself (Table 1), suggesting that, unlike B. subtilis, the viability of the aPBP-less strain or any other PG synthase mutant of Cglu does not depend on a transcriptional response that enhances the expression of other PG synthases.
TABLE 1.
Expression of genes encoding PG synthases in mutant backgrounds
| Gene | Normalized gene expression relative to WT in strains with mutation of:a |
||||
|---|---|---|---|---|---|
| ΔponA | ΔponB | ΔrodA | ΔponA ΔponB | ΔponA ΔrodA | |
| ponA | 0.001 | 1.010 | 1.096 | 0.001 | 0.001 |
| ponB | 1.023 | 0.001 | 1.119 | 0.001 | 1.145 |
| rodA | 0.993 | 1.022 | 0.001 | 1.135 | 0.002 |
| ftsW | 1.102 | 1.047 | 1.040 | 1.143 | 1.183 |
| pbpA/pbp2b | 1.042 | 1.028 | 1.367 | 1.145 | 1.575 |
| ftsI | 1.078 | 1.007 | 0.985 | 1.287 | 1.247 |
| pbp/pbp2a | 1.188 | 1.277 | 1.241 | 1.287 | 1.549 |
Fold change of normalized read count for each of the listed genes in each deletion mutant relative to the read count from wild-type (MB001) cells. Raw read counts for each gene were normalized across samples to account for sample coverage. Tabulated data represent the average of duplicate RNA-seq runs for each strain.
The different types of PG synthases have distinct polar localization patterns.
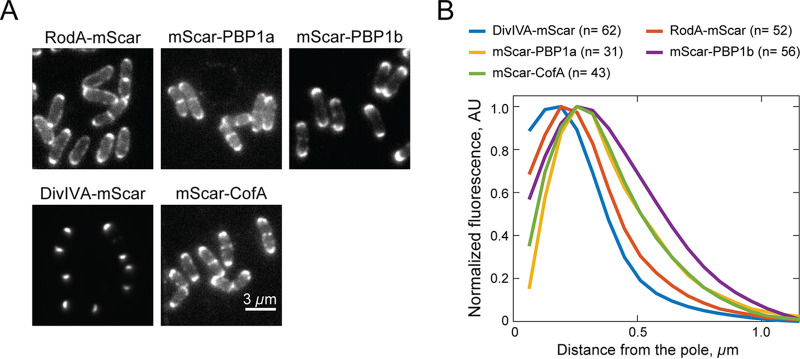
During dispersed elongation in B. subtilis and E. coli, the RodA-PBP2 and the aPBP synthases display different dynamic localizations (7), suggesting that they are performing different roles in the growth of the cylindrical cell body. We were therefore curious whether RodA and the aPBPs might have discernably different localization patterns at the cell pole of Cglu. Strains producing mScar fusions to RodA, PBP1a, or PBP1b as the sole copy of the corresponding synthase were constructed. All three fusion proteins were deemed functional, as they complemented the drug or heat sensitivity phenotypes of mutants deleted for the gene encoding the relevant native synthase (Fig. S1). Consistent with previous reports (30, 33), cells producing these fusion proteins all displayed prominent fluorescent signals at the cell poles and at sites of cell division (Fig. 6A). Notably, upon close examination, the patterns generated by mScar-RodA versus that of the tagged aPBPs appeared to be subtly different. The RodA-mScar signal was more concentrated at the cell tip, whereas the signals for the aPBPs appeared to encompass a greater area of the cell pole (Fig. 6A). Quantification of the fluorescence distributions along the long axis of the cell extending from the brightest pole confirmed the difference in localization patterns between RodA and the aPBPs (Fig. 6B). Notably, the distribution of RodA-mScar mirrored the tight localization of DivIVA-mScar at the polar tip, whereas the localization pattern of an mScar fusion to CofA, a PBP1a interaction partner (37), displayed a broader localization pattern similar to that of the aPBPs (Fig. 6A and B).
FIG 6.
Spatial distribution of RodA and the aPBPs at the cell poles. (A) Representative fluorescence micrographs of Cglu cells producing the indicated mScar fusions as the sole copy of the corresponding genes either from the native locus (DivIVA-mScar, strain HL23) or integrated plasmids pJSW19 (mScar-PBP1a) in strain HL37 (ΔponA), pJSW18 (mScar-CofA) in strain JS8 (ΔcofA), pJWS94 (mScar-PBP1b) in strain JS20 (ΔponB), or pJWS33 (mScar-RodA) in strain HL31 (ΔrodA). Overnight cultures grown in BHI medium at 30°C were diluted 1:1,000 in BHI medium supplemented with 0.3 mM theophylline and grown at 30°C. When the OD600 reached 0.2 to 0.3, cells were diluted 10-fold and loaded into a CellASIC microfluidic device for imaging by fluorescence microscopy. (B) Quantification of polar fluorescence distributions of the indicated mScar fusions. Following cell segmentation by Oufti (48), a MATLAB-based script was used to identify the brightest pole of each cell, aligning the cells by setting the tip of the brightest pole to position zero. The average fluorescence intensity distribution across all cells as a function of distance from the pole was then quantified. The fluorescence profiles shown were plotted following background subtraction.
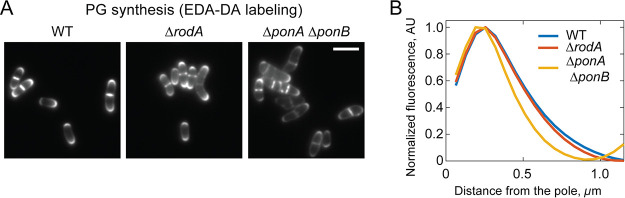
To determine if the differential spatial distribution of the PG synthases might be reflected in the pattern of PG biogenesis, we monitored the synthesis of new PG in mutants relying on either RodA or PBP1b alone for polar growth using the d-Ala-d-Ala dipeptide analog EDA-DA (40). This chemical probe is incorporated into the peptide stem of PG precursors, which are then used in the construction of the PG layer. The spatial localization of probe incorporation into the cell wall can then be assessed in fixed cells using click chemistry to couple a fluorescent dye to the EDA moiety. After a 5-min pulse with EDA-DA, ΔrodA cells, which rely on the aPBPs for polar growth, were found to incorporate new PG over a broader portion of the cell pole than ΔponA ΔponB cells, which use RodA for elongation (Fig. 7A and B). Thus, the patterns of new PG incorporation in these strains correlate well with the corresponding polar distributions observed for the type of enzyme performing the synthesis. Although more advanced microscopic methods will be required to gain a more definitive higher-resolution picture of polar PG synthesis, these results suggest that the aPBP and SEDS-bPBP synthases involved in apical elongation may have specialized roles in the process and operate from distinct polar subregions.
FIG 7.
Localization of PG insertion at the poles of cells inactivated for subsets of PG synthases. (A) Representative fluorescent micrographs of MB001 (WT) and its indicated derivatives HL31 (ΔrodA) and JS36 (ΔponA ΔponB) showing incorporation of nascent PG over a period of 5 min. Overnight cultures grown in BHIS medium at 30°C were diluted 1:1,000 in BHIS medium, grown to an OD600 of 0.2, gently pelleted, and resuspend into BHIS medium supplemented with 0.5 mM d-Ala-d-ala dipeptide derivative EDA-DA. Cells were fixed with 70% ethanol after 5 min, followed by click chemistry labeling of the EDA moiety with Alexa 488. (B) Quantification of polar PG labeling was performed as described for Fig. 6B.
DISCUSSION
In this report, we revisited the requirements of different PG synthases for polar growth in Cglu. Our results revealed a surprising flexibility for this mode of cell elongation and morphogenesis. Unlike the dispersed mode of PG synthesis employed by many model rod-shaped bacteria, which has a strict requirement for the SEDS-type PG polymerase RodA, polar growth and rod-shape determination in Cglu remains functional when either RodA or the aPBPs are inactivated. Although we cannot rule out a contribution of the SEDS-type PG polymerase FtsW to cell elongation in cells inactivated for RodA, we think this possibility is unlikely, because: (i) the overproduction of FtsW-green fluorescent protein (GFP) in Cglu was previously shown not to correct the growth defect of ΔrodA cells (30), (ii) FtsW is dispensable for cell elongation in Streptomyces coelicolor (41, 42), and (iii) FtsW was found to localize exclusively to cell division sites in Cglu (30). Also, in all other organisms in which it has been studied, FtsW has been shown to specifically function in cell division. Thus, our genetic and cytological analyses indicate that there is inherent plasticity in the mechanism of PG biogenesis during polar elongation that allows the process to continue in Cglu with either one of the two major types of PG synthase enzymes.
Potential division of labor between RodA and the aPBPs during polar elongation.
Despite the functionality of polar elongation in Cglu cells lacking either RodA or the aPBPs, the mutants have a slow-growth phenotype, indicating that both types of synthases are required for optimal fitness. The different types of enzymes are therefore unlikely to serve completely redundant functions in polar PG biogenesis. Some specialization is probable. Accordingly, RodA and the aPBPs appear to have distinct localization patterns at the cell poles in Cglu, with RodA being more concentrated at the cell tip and the aPBPs occupying a broader distribution around the pole. Notably, PonA1 in Msmeg, which is the ortholog of PBP1b in Cglu, has also been found to have a broad distribution extending from the fast-growing old cell pole (32). Moreover, the dispensability of RodA and essentiality of PonA1 for Msmeg growth (31, 34, 35) provide additional support for the two types of synthases having at least some specific roles during polar PG biogenesis. What these differential roles are and whether the aPBPs function to fortify a foundational PG structure deposited by RodA-PBP2b at the pole similar to their proposed role in the dispersed mode of cell elongation will require further investigation. Nevertheless, the fact that Cglu can elongate without the aPBPs indicates that, in contrast to current thinking, the process of polar elongation does not fundamentally require their activity.
Connection between cell elongation and daughter cell separation.
Cells inactivated for RodA or the aPBPs were found to be elongated and to contain multiple septa relative to wild-type cells. A similar phenotype was previously observed for mutants lacking PBP2b (33), the presumed partner of RodA in the elongation SEDS-bPBP synthase. This observation was surprising given that these cell wall synthases are thought to function predominantly in the polar elongation process. However, the cytological experiments reported here and in previous studies (30, 33) indicate that all of these factors localize at division sites as well as the cell poles. This division site recruitment of the cell elongation PG synthases coupled with the division phenotype displayed by the mutants suggests that components of the elongation machinery may actively promote the daughter cell separation process. Accordingly, PonA1 in Mtb has been found to interact with the PG hydrolase RipA that is involved in cell separation (31). Although it is possible that RodA or the aPBPs function as part of the divisome, another attractive possibility that warrants further investigation is that it is not just PG cleavage at the septum (43) but also the activation of PG synthesis by the elongation enzymes at division sites that provides the mechanical strain necessary to promote the extremely rapid septal cracking observed as corynebacterial and mycobacterial cells separate (44).
Regulation of ponB expression.
Not much is known about the transcriptional regulation of genes encoding aPBPs in any organism. Our genetic analysis of ΔponA ΔrodA cells identified suppressors that promote growth of the mutant by increasing ponB expression. These suppressors were found to contain one of three different changes in the chromosomal region just upstream of ponB, two of which were found to enhance its transcription. The mechanism by which these changes elicit this effect is not clear, but the observation that they only function in cis suggests that they may function by altering the sequence of a local promoter to strengthen it, potentially by modulating the binding of a transcription factor, or by modifying the structure and/or stability of the ponB mRNA. Further work will be required to differentiate between these possibilities and determine whether a stress response or a related type of regulatory system might be responsible for the underlying phenomenon.
Conclusion.
One of the first steps toward elucidating the mechanism of a process is determining what the minimal required components are. Here, we made the surprising finding that either one of the two known types of PG synthases can effectively promote polar growth of Cglu. Therefore, unlike dispersed cell elongation and cell division, which have an absolute requirement for a SEDS-bPBP-type synthase, the underlying mechanism promoting apical growth in Corynebacterineae must be inherently adaptable such that at least in some members of this suborder either a SEDS-bPBP or an aPBP synthase can produce a form of PG capable of extending the pole and elongating the cell. A challenge for the future will be to identify the factors that contribute to the flexibility of this growth mode and whether it makes the process more resilient to stress or antibiotic challenge than other mechanisms, perhaps as a means to enhance the survival of organisms that grow apically in the competitive soil environment.
MATERIALS AND METHODS
Media, bacterial strains, and plasmids.
All Cglu strains used in the reported experiments are derivatives of MB001 (45). Strains were grown in brain heart infusion (BHI) medium (BD) that was supplemented with 91 g/liter of sorbitol (called BHIS) (Sigma) where indicated. When necessary, 15 μg/ml kanamycin (Kan) or 2.5 μg/ml apramycin (Apra) was added to the medium to select for plasmids/expression constructs. Theophylline was added where indicated at 0.3 mM unless otherwise specified. Whenever necessary, antibiotics for E. coli cultures were used at 25 μg/ml (Kan) or 50 μg/ml (Apra). Details for plasmid constructions are provided in Text S1 in the supplemental material. All strains and plasmids used are listed in Table S1 and S2, respectively.
Supplemental text with plasmid construction procedures. Download Text S1, DOCX file, 0.02 MB (22.6KB, docx) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains used in this study. Download Table S1, DOCX file, 0.01 MB (15KB, docx) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download Table S2, DOCX file, 0.01 MB (15.1KB, docx) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strain construction.
For gene deletion in Cglu, we used the pCRD206 temperature-sensitive plasmid (46). Briefly, the deletion allele with regions corresponding to approximately 750 bp upstream and downstream of the desired deletion were inserted into pCRD206. The resulting plasmid was transformed into the appropriate recipient strain. Transformants were selected and propagated on BHIS agar supplemented with Kan at 25°C. To select for clones, colonies were purified on a BHI Kan plate and incubated at 30°C for 36 h. The resulting Kanr colonies were grown in BHI liquid medium at 25°C overnight. The culture was then spread on a BHIS agarose plate supplemented with 10% sucrose and grown at 30°C. The resulting colonies were replica patched on BHIS and BHIS Kan plates to identify Kan-sensitive colonies. The deletion allele was finally confirmed by diagnostic colony PCR. The above-described procedure was modified if the strain had an inducible complementation construct by adding 0.3 mM theophylline to all of the plates to induce expression of the cloned gene. Plasmid integration at the attB1 site was performed using derivatives of the pK-PIM vector (47). Plasmids containing the desired expression construct were introduced into the recipient by direct transformation.
Preparation of electrocompetent Cglu.
A stationary-phase Cglu culture (10 ml) was diluted into 1 liter of BHIS medium that was supplemented with 25 g glycine, 0.4 g isoniazid, and 0.1% Tween 80 and incubated with shaking at 18°C. The culture was chilled on ice for 1 h after the optical density at 600 nm (OD600) reached 0.5 (typically in 16 to 18 h). Cells were then collected by centrifugation at 4,000 × g for 20 min. The pellet was washed once with 500 ml chilled 10% glycerol and three additional times with 100 ml chilled 10% glycerol. Cell density was adjusted to an OD600 of 20 before use for electroporation.
Electroporation of DNA.
Approximately 100 ng of DNA was mixed with 100 μl of electrocompetent cells in a 1-mm electroporation cuvette (Genesee Scientific). The cells were then electroporated at 1.7 kV using a MicroPulser electroporator (Bio-Rad). The cells were recovered in 1 ml BHIS medium and then immediately heat shocked for 6 min at 46°. The cells were then grown at 30° for 1 h before plating on the appropriate agar medium.
Microscopy.
Growth conditions and staining procedures prior to microscopy are described in the figure legends. Images were cropped and adjusted using FIJI software. Measurements of cell lengths and fluorescence signals at the single-cell level were carried out using Oufti (48).
For experiments that utilized the CellASICs device, cells were loaded into the CellASIC Onix B04 microfluidic plates (Millipore Sigma) that were attached to the microscope by using a multiwell insert. Each imaging chamber was flushed with the appropriate growth medium before loading the cells. During the course of the time-lapse imaging, appropriate growth media were supplied to the cells using a constant pressure of 60 lb/in2. To avoid reduction of growth rate due to phototoxicity, we lowered the intensity of the excitation light using neutral density filters (at least ND8) in all experiments.
Images were obtained using a Nikon Ti inverted microscope that is fitted with a Nikon motorized stage with an OkoLab gas incubator with a slide insert attachment, an Andor Zyla 4.2 Plus scientific complementary metal-oxide semiconductor (sCMOS) camera, Lumencore SpectraX light-emitting diode (LED) illumination, Plan Apo lambda 100×/1.45 Oil Ph3 DM lens objective, and Nikon Elements 4.30 acquisition software. Images in the green and red channels were taken using Chroma 49002 and 49008 filter cubes, respectively. The microscope was maintained at 30°C using a custom-made environmental control chamber.
RNA sequencing.
Overnight cultures of each of the strains were diluted 1:200 in BHIS medium and allowed to grow to an OD600 of 0.3. An aliquot of cells (1 ml) was then pelleted and frozen at −80°C. Library preparation and Illumina sequencing were performed by GENEWIZ (South Plainfield, NJ, USA). Data analysis was performed by CLC genomics workbench.
Bocillin labeling.
Bocillin labeling was performed as described previously (37).
Beta-galactosidase assay.
The above-indicated strains were grown to an OD600 of 0.3. Cells from 1 ml of culture were then pelleted and resuspended in 1 ml Z buffer (Na2HPO4-NaH2PO4 [pH 7.0], 10 mM KCl, 1 mM MgSO4, 50 mM beta-mercaptoethanol) with 2% toluene to permeabilize the cells. Beta-galactosidase activity was determined with permeabilized cells as described by Miller (49).
Suppressor selection.
Eight independent colonies of the ΔrodA ΔponA strain were grown overnight in BHIS medium. The cultures were normalized to an OD600 of 0.5, and 1-μl or 2-μl aliquots were plated on BHI agar, resulting in 0 to 22 suppressors on each plate. Two to three suppressors from each of the selection plates were then selected for the whole-genome sequencing. Whole-genome sequencing of the suppressors was performed as described previously (9).
ACKNOWLEDGMENTS
We thank all members of the Bernhardt and Rudner labs for advice and helpful discussions as well as the microscopy services provided by Paula Montero Llopis and her team at the Microscopy Resources on the North Quad (MicRoN) core facility at HMS.
This work was supported by the National Institutes of Health (AI083365 to T.G.B), and Investigator funds from the Howard Hughes Medical Institute. H.C.L. was supported in part by the Life Sciences Research Foundation, where he was a Simons Fellow. J.W.S. was supported in part by the T32 Bacteriology PhD Training Program (AI132120-02) awarded to the Harvard Graduate Program in Bacteriology.
Footnotes
Citation Sher JW, Lim HC, Bernhardt TG. 2021. Polar growth in Corynebacterium glutamicum has a flexible cell wall synthase requirement. mBio 12:e00682-21. https://doi.org/10.1128/mBio.00682-21.
Contributor Information
Thomas G. Bernhardt, Email: thomas_bernhardt@hms.harvard.edu.
M. Sloan Siegrist, University of Massachusetts Amherst
REFERENCES
- 1.Egan AJF, Errington J, Vollmer W. 2020. Regulation of peptidoglycan synthesis and remodelling. Nat Rev Microbiol 18:446–460. doi: 10.1038/s41579-020-0366-3. [DOI] [PubMed] [Google Scholar]
- 2.Ishino F, Mitsui K, Tamaki S, Matsuhashi M. 1980. Dual enzyme activities of cell wall peptidoglycan synthesis, peptidoglycan transglycosylase and penicillin-sensitive transpeptidase, in purified preparations of Escherichia coli penicillin-binding protein 1A. Biochem Biophys Res Commun 97:287–293. doi: 10.1016/S0006-291X(80)80166-5. [DOI] [PubMed] [Google Scholar]
- 3.Nakagawa J, Tamaki S, Tomioka S, Matsuhashi M. 1984. Functional biosynthesis of cell wall peptidoglycan by polymorphic bifunctional polypeptides. Penicillin-binding protein 1Bs of Escherichia coli with activities of transglycosylase and transpeptidase. J Biol Chem 259:13937–13946. doi: 10.1016/S0021-9258(18)89835-0. [DOI] [PubMed] [Google Scholar]
- 4.Ye XY, Lo MC, Brunner L, Walker D, Kahne D, Walker S. 2001. Better substrates for bacterial transglycosylases. J Am Chem Soc 123:3155–3156. doi: 10.1021/ja010028q. [DOI] [PubMed] [Google Scholar]
- 5.Bertsche U, Breukink E, Kast T, Vollmer W. 2005. In vitro murein peptidoglycan synthesis by dimers of the bifunctional transglycosylase-transpeptidase PBP1B from Escherichia coli. J Biol Chem 280:38096–38101. doi: 10.1074/jbc.M508646200. [DOI] [PubMed] [Google Scholar]
- 6.Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, Rudner DZ. 2016. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PDA, Suh H, Marto JA, Garner EC, Bernhardt TG. 2016. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol 1:16172. doi: 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taguchi A, Welsh MA, Marmont LS, Lee W, Sjodt M, Kruse AC, Kahne D, Bernhardt TG, Walker S. 2019. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat Microbiol 4:587–594. doi: 10.1038/s41564-018-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohs PDA, Buss J, Sim SI, Squyres GR, Srisuknimit V, Smith M, Cho H, Sjodt M, Kruse AC, Garner EC, Walker S, Kahne DE, Bernhardt TG. 2018. A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genet 14:e1007726. doi: 10.1371/journal.pgen.1007726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sjodt M, Rohs PDA, Gilman MSA, Erlandson SC, Zheng S, Green AG, Brock KP, Taguchi A, Kahne D, Walker S, Marks DS, Rudner DZ, Bernhardt TG, Kruse AC. 2020. Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex. Nat Microbiol 5:813–820. doi: 10.1038/s41564-020-0687-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel RA, Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 12.Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. 2011. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domínguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Söldner R, Carballido-López R. 2011. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 14.van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. 2011. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci U S A 108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wachi M, Doi M, Okada Y, Matsuhashi M. 1989. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J Bacteriol 171:6511–6516. doi: 10.1128/JB.171.12.6511-6516.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kruse T, Bork-Jensen J, Gerdes K. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol 55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- 17.Bendezú FO, Hale CA, Bernhardt TG, de Boer PAJ. 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacobs-Wagner C. 2009. RodZ, a component of the bacterial core morphogenic apparatus. Proc Natl Acad Sci U S A 106:1239–1244. doi: 10.1073/pnas.0810794106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiomi D, Sakai M, Niki H. 2008. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J 27:3081–3091. doi: 10.1038/emboj.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bratton BP, Shaevitz JW, Gitai Z, Morgenstein RM. 2018. MreB polymers and curvature localization are enhanced by RodZ and predict E. coli’s cylindrical uniformity. Nat Commun 9:2797. doi: 10.1038/s41467-018-05186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bendezú FO, de Boer PAJ. 2008. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol 190:1792–1811. doi: 10.1128/JB.01322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vigouroux A, Cordier B, Aristov A, Alvarez L, Özbaykal G, Chaze T, Oldewurtel ER, Matondo M, Cava F, Bikard D, van Teeffelen S. 2020. Class-A penicillin binding proteins do not contribute to cell shape but repair cell-wall defects. Elife 9:e51998. doi: 10.7554/eLife.51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McPherson DC, Popham DL. 2003. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J Bacteriol 185:1423–1431. doi: 10.1128/JB.185.4.1423-1431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dion MF, Kapoor M, Sun Y, Wilson S, Ryan J, Vigouroux A, van Teeffelen S, Oldenbourg R, Garner EC. 2019. Bacillus subtilis cell diameter is determined by the opposing actions of two distinct cell wall synthetic systems. Nat Microbiol 4:1294–1305. doi: 10.1038/s41564-019-0439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figueroa-Cuilan WM, Brown PJB. 2018. Cell wall biogenesis during elongation and division in the plant pathogen Agrobacterium tumefaciens, p 87–110. In Gelvin SB (ed), Agrobacterium biology, from basic science to biotechnology. Springer International Publishing, New York, NY. [DOI] [PubMed] [Google Scholar]
- 26.Baranowski C, Rego EH, Rubin EJ. 2019. The dream of a Mycobacterium. Microbiol Spectr 7:GPP3-0008-2018. doi: 10.1128/microbiolspec.GPP3-0008-2018. [DOI] [PubMed] [Google Scholar]
- 27.Kieser KJ, Rubin EJ. 2014. How sisters grow apart: mycobacterial growth and division. Nat Rev Microbiol 12:550–562. doi: 10.1038/nrmicro3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hempel AM, Wang S, Letek M, Gil JA, Flärdh K. 2008. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J Bacteriol 190:7579–7583. doi: 10.1128/JB.00839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letek M, Ordóñez E, Vaquera J, Margolin W, Flärdh K, Mateos LM, Gil JA. 2008. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J Bacteriol 190:3283–3292. doi: 10.1128/JB.01934-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sieger B, Schubert K, Donovan C, Bramkamp M. 2013. The lipid II flippase RodA determines morphology and growth in Corynebacterium glutamicum. Mol Microbiol 90:966–982. doi: 10.1111/mmi.12411. [DOI] [PubMed] [Google Scholar]
- 31.Hett EC, Chao MC, Rubin EJ. 2010. Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria. PLoS Pathog 6:e1001020. doi: 10.1371/journal.ppat.1001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baranowski C, Welsh MA, Sham L-T, Eskandarian HA, Lim HC, Kieser KJ, Wagner JC, McKinney J, Fantner GE, Ioerger TR, Walker S, Bernhardt TG, Rubin EJ, Rego EH. 2018. Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. Elife 7:e37516. doi: 10.7554/eLife.37516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valbuena N, Letek M, Ordóñez E, Ayala J, Daniel RA, Gil JA, Mateos LM. 2007. Characterization of HMW-PBPs from the rod-shaped actinomycete Corynebacterium glutamicum: peptidoglycan synthesis in cells lacking actin-like cytoskeletal structures. Mol Microbiol 66:643–657. doi: 10.1111/j.1365-2958.2007.05943.x. [DOI] [PubMed] [Google Scholar]
- 34.Arora D, Chawla Y, Malakar B, Singh A, Nandicoori VK. 2018. The transpeptidase PbpA and noncanonical transglycosylase RodA of Mycobacterium tuberculosis play important roles in regulating bacterial cell lengths. J Biol Chem 293:6497–6516. doi: 10.1074/jbc.M117.811190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kieser KJ, Boutte CC, Kester JC, Baer CE, Barczak AK, Meniche X, Chao MC, Rego EH, Sassetti CM, Fortune SM, Rubin EJ. 2015. Phosphorylation of the peptidoglycan synthase PonA1 governs the rate of polar elongation in mycobacteria. PLoS Pathog 11:e1005010. doi: 10.1371/journal.ppat.1005010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kieser KJ, Baranowski C, Chao MC, Long JE, Sassetti CM, Waldor MK, Sacchettini JC, Ioerger TR, Rubin EJ. 2015. Peptidoglycan synthesis in Mycobacterium tuberculosis is organized into networks with varying drug susceptibility. Proc Natl Acad Sci U S A 112:13087–13092. doi: 10.1073/pnas.1514135112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sher JW, Lim HC, Bernhardt TG. 2020. Global phenotypic profiling identifies a conserved actinobacterial cofactor for a bifunctional PBP-type cell wall synthase. Elife 9:e54761. doi: 10.7554/eLife.54761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, Gadella TWJ. 2017. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods 14:53–56. doi: 10.1038/nmeth.4074. [DOI] [PubMed] [Google Scholar]
- 39.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, VanNieuwenhze MS. 2012. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew Chem Int Ed Engl 51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, Maurelli AT. 2014. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506:507–510. doi: 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennett JA, Yarnall J, Cadwallader AB, Kuennen R, Bidey P, Stadelmaier B, McCormick JR. 2009. Medium-dependent phenotypes of Streptomyces coelicolor with mutations in ftsI or ftsW. J Bacteriol 191:661–664. doi: 10.1128/JB.01048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mistry BV, Sol RD, Wright C, Findlay K, Dyson P. 2008. FtsW is a dispensable cell division protein required for Z-ring stabilization during sporulation septation in Streptomyces coelicolor. J Bacteriol 190:5555–5566. doi: 10.1128/JB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim HC, Sher JW, Rodriguez-Rivera FP, Fumeaux C, Bertozzi CR, Bernhardt TG. 2019. Identification of new components of the RipC-FtsEX cell separation pathway of Corynebacterineae. PLoS Genet 15:e1008284. doi: 10.1371/journal.pgen.1008284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou X, Rodriguez-Rivera FP, Lim HC, Bell JC, Bernhardt TG, Bertozzi CR, Theriot JA. 2019. Sequential assembly of the septal cell envelope prior to V snapping in Corynebacterium glutamicum. Nat Chem Biol 15:221–231. doi: 10.1038/s41589-018-0206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baumgart M, Unthan S, Rückert C, Sivalingam J, Grünberger A, Kalinowski J, Bott M, Noack S, Frunzke J. 2013. Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl Environ Microbiol 79:6006–6015. doi: 10.1128/AEM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okibe N, Suzuki N, Inui M, Yukawa H. 2011. Efficient markerless gene replacement in Corynebacterium glutamicum using a new temperature-sensitive plasmid. J Microbiol Methods 85:155–163. doi: 10.1016/j.mimet.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 47.Oram M, Woolston JE, Jacobson AD, Holmes RK, Oram DM. 2007. Bacteriophage-based vectors for site-specific insertion of DNA in the chromosome of corynebacteria. Gene 391:53–62. doi: 10.1016/j.gene.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paintdakhi A, Parry B, Campos M, Irnov I, Elf J, Surovtsev I, Jacobs-Wagner C. 2016. Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol Microbiol 99:767–777. doi: 10.1111/mmi.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 50.Desai SK, Gallivan JP. 2004. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J Am Chem Soc 126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- 51.Lord SJ, Velle KB, Mullins RD, Fritz-Laylin LK. 2020. SuperPlots: communicating reproducibility and variability in cell biology. J Cell Biol 219:e202001064. doi: 10.1083/jcb.202001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complementation of PG synthase mutant phenotypes. (A to C) Overnight cultures of MB001 (WT) and the indicated derivatives HL18 (ΔponA), JS20 (ΔponB), and HL31 (ΔrodA) grown in BHIS medium at 30°C were normalized to an OD600 of 0.5, serially diluted 10-fold, and spotted (5 μl) onto BHI or BHIS agar medium supplemented with theophylline (0.3 mM), ampicillin (0.2 μg/ml), or meropenem (0.03 μg/ml) where indicated. Plates were incubated for 24 h at 30°C and photographed. Plasmids used were pJWS19 (Psod::riboE1-mScar-ponA Kanr) and pJWS116 (Psod::riboE1-mScar-ponA Aprar] (A), pJWS94 (Psod::riboE1-mScar-ponB Kanr) and pJWS117 (Psod::riboE1-mScar-ponB Aprar) (B), and pJWS33 (Psod::riboE1-rodA-mScar Kanr) (C). Download FIG S1, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cell division phenotype of ΔrodA cells and complementation by mScar-RodA. (A) Representative phase contrast micrographs of MB001 (WT) and its HL31 (ΔrodA) derivative. Overnight cultures grown in BHIS medium at 30°C were diluted 1:1,000 in BHI medium and grown at 30°C. When the OD600 reached 0.2 to 0.3, cells were diluted 10-fold and loaded into a CellASIC microfluidic device and imaged by phase-contrast microscopy. Where indicated, RodA-mScar production was induced from integrated pJWS33 (Psod::riboE1-rodA-mScar Kanr) with theophylline (0.3 μM). The yellow arrowhead indicates longer cells within the population, while cyan arrowheads indicate unusually small cells that originated from recently v-snapped long cells. (B) Representative fluorescence micrographs of the strains from panel A grown as described above but pulsed labeled with TADA for 5 min prior to imaging. (C) Histograms showing cell length distributions of the ΔrodA mutant with or without RodA-mScar production. Download FIG S2, TIF file, 1.0 MB (985.8KB, tif) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Morphology of ΔponA ΔponB cells grown in BHI medium without sorbitol. An overnight culture of JS36 (ΔponA ΔponB) grown in BHIS medium at 30°C was diluted 1:1,000 in BHI medium and grown at 30°C. When the OD600 reached 0.1, cells were loaded on BHI agar pads and imaged by phase-contrast microscopy. Download FIG S3, TIF file, 0.6 MB (659.8KB, tif) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bocillin-labeling controls for determining PBP identity. Cells of MB001 (WT) and its derivatives HL18 (ΔponA), JS20 (ΔponB), JS12 (Δpbp2a), and HL37 (Δpbp2b) labeled with Bocillin-FL. Membrane extracts were then prepared from the labeled cells, and 5 μg of total protein for each was subjected to SDS-PAGE. Bocillin-FL-labeled proteins were then detected using a Typhoon 9500 imager with excitation at 488 nm and emission at 530 nm. (A) Gel was run at 110 V for 1.5 h. (B) Gel was run at 110 V for 3 h to help resolve the multiple bands at running at ∼100 kDa. Download FIG S4, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Morphology of cells with PBP1b as their only elongation PG synthase. Overnight cultures of MB001 (WT) and the indicated derivatives of JS38 (ΔponA ΔrodA) grown in BHIS medium at 30°C were diluted 1:1,000. The cultures were then grown to an OD600 of ∼0.05 before being loaded into CellASIC devices for imaging by phase-contrast microscopy. Cell length and width were measured for a population of cells of each strain (n > 240) using Oufti. Representative micrographs of each strain are shown adjacent to plots of the corresponding morphological measurements. Each dot in the plots represents measurements from a single cell. The lines indicate the mean measurement plus or minus one standard deviation. The medium was supplemented with theophylline (0.6 mM) to induce mScar-PBP1b production in the pJWS117-containing strain. Download FIG S5, TIF file, 2.5 MB (2.5MB, tif) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental text with plasmid construction procedures. Download Text S1, DOCX file, 0.02 MB (22.6KB, docx) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains used in this study. Download Table S1, DOCX file, 0.01 MB (15KB, docx) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download Table S2, DOCX file, 0.01 MB (15.1KB, docx) .
Copyright © 2021 Sher et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.