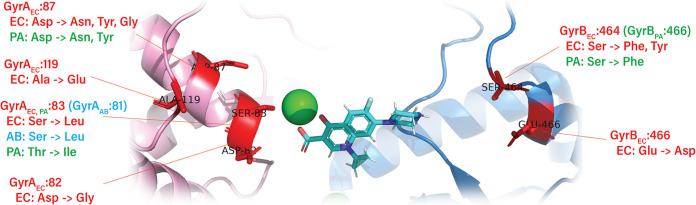
FIG 3.
Amino acid substitutions in GyrA (pink chain)/GyrB (blue chain) observed during the course of the morbidostat-based experimental evolution of CIP resistance in E. coli (EC; red text), A. baumannii (AB; blue text), and P. aeruginosa (PA; green text) mapped on a three-dimensional structure (PDB accession number 6RKW). The ciprofloxacin molecule (blue) and Mg2+ ion (green) were added by structural alignment of 6RKW with the structure of Mycobacterium tuberculosis gyrase bound to CIP (PDB accession number 5BTC). The substitution equivalent to P. aeruginosa GyrB:Leu128Pro is not shown. It is located near the ATP-binding site of GyrB. Chain A of 6RKW was aligned to chain A of 5BTC using FATCAT. The same rotation translation was then applied to all chains in 6RKW to align the full structure.