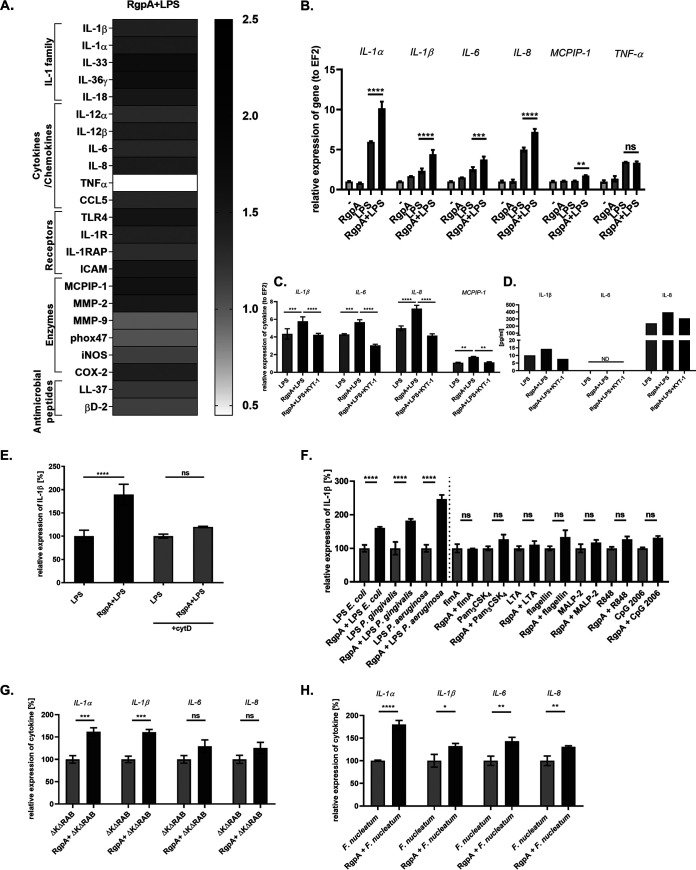
FIG 4.
RgpA induces hyperresponsiveness of gingival keratinocytes to endotoxin and Gram-negative bacteria. (A) TIGK cells were prestimulated with 2 nM RgpA for 1 h and treated with lipopolysaccharide (LPS; 20 μg/ml) or infected with bacteria for an additional 3 h. Heat map represents the changes in the expression of genes regulated by LPS signaling in the presence of RgpA. Relative mRNA levels were determined by quantitative reverse transcription-PCR (qRT-PCR). Expression of each gene in cells stimulated only with LPS was established as 1. (B) Quantitative RT-PCR analysis of the proinflammatory cytokines and MCPIP-1 mRNA level after treatment of cells with RgpA, LPS, and/or RgpA 1 h prior to LPS in comparison to untreated cells. (C, D) Relative expression of IL-1β, IL-6, IL-8 and MCPIP-1 at the mRNA and protein level in TIGK cells. Gingival keratinocytes were pretreated for 1 h with RgpA, and then the specific-RgpA inhibitor KYT-1 was added for 30 min. Subsequently, cells were stimulated with LPS for 3 h. ND, not detected. (E) Relative expression of IL-1β after treatment of TIGK cells with cytochalasin D (+cytD) for 30 min prior to their stimulation with LPS and RgpA/LPS in comparison to cytochalasin D-untreated cells. LPS-stimulated cells are presented as 100%. (F) Comparison of IL-1β mRNA expression determined by qRT-PCR after a 3-h reaction of TIGKs to different Toll-like receptor (TLR) agonists in comparison to cells additionally pretreated with RgpA for 1 h. TIGKs stimulated with corresponding agonists are presented as 100%. (G, H) Gingiva keratinocytes were preincubated for 1 h with RgpA (2 nM) and then exposed for 3 h to (G) the P. gingivalis gingipain-deficient ΔKΔRAB strain at an MOI of 1:25, or (H) F. nucleatum at an MOI of 1:10. The level of proinflammatory cytokines was determined by qRT-PCR. TIGKs stimulated with bacteria are presented as 100%. Data represent mean values from three independent experiments ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.