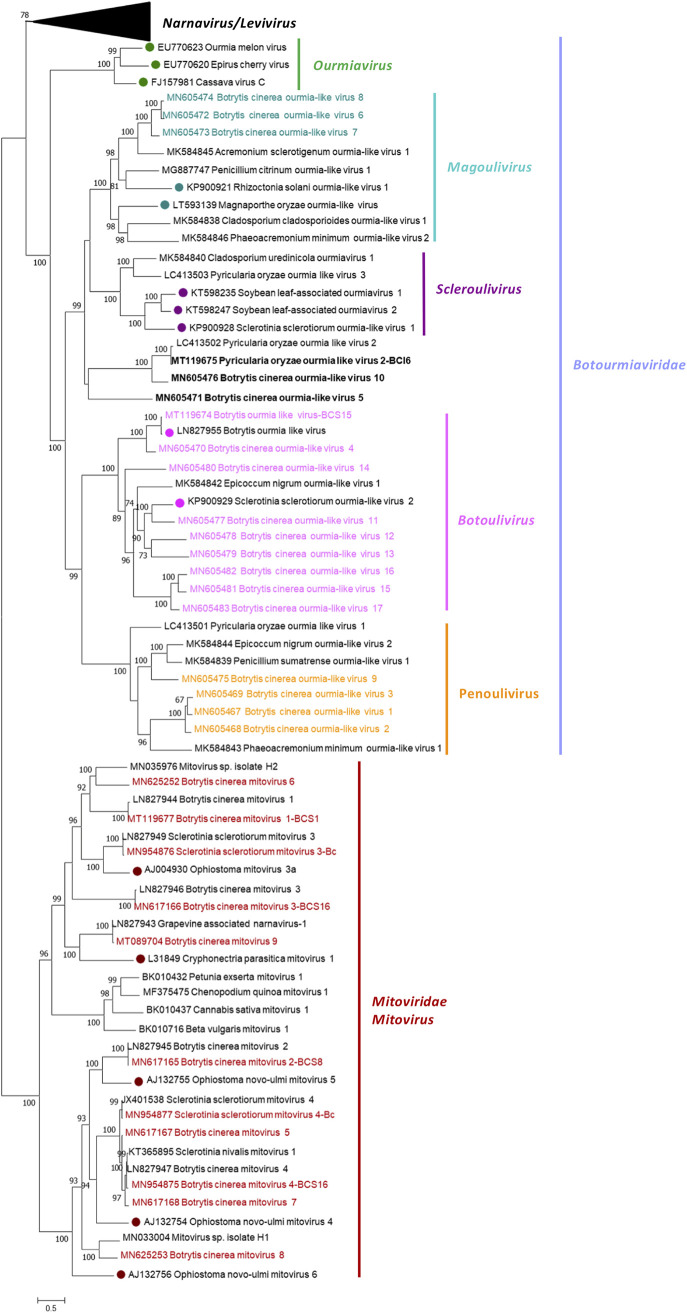
FIG 4.
Mitovirus and botourmiavirus phylogenetic tree. A phylogenetic tree was computed by using the IQ-TREE stochastic algorithm to infer phylogenetic trees by maximum likelihood (model of substitution: VT+F+I+G4). A consensus tree was constructed from 1,000 bootstrap trees (log likelihood of consensus tree, –126059.306862). The branch of the narnaviruses and leviviruses is collapsed. All bootstrap values (%) of >65 are represented at each node of the tree. Branch lengths are proportional to the number of amino acid substitutions and are measured by a scale bar.