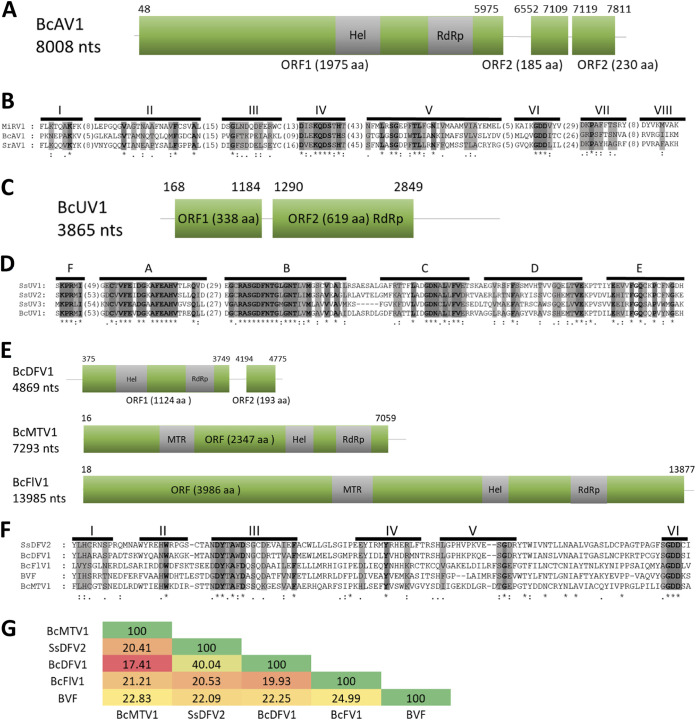
FIG 6.
Sequence properties of Botrytis cinerea ssRNA+ mycoviruses. The conserved motifs of the RdRps are shaded with light and dark gray colors. Asterisks indicate identical amino acid residues, and colons indicate similar residues. Amino acids in parentheses show the positions of amino acid residues that are not listed. (A) Schematic representation of Botrytis cinerea alpha-like virus 1 (BcAV1) RNA genome showing location of ORFs. (B) Amino acid sequence alignment of alphavirus RdRps (I to VIII), with the reference genomes Morchella importuna RNA virus 1 (MiRV1, MK279480.1) and Sclerotium rolfsii alphavirus-like virus 1 (SrAV1, MH766488.1). (C) Schematic representation of Botrytis cinerea umbra-like virus 1 (BcUV1) RNA genomes showing the locations of ORFs. (D) Amino acid sequence alignment of umbravirus RdRps showing conserved motifs F to E, with the reference genome Sclerotinia sclerotiorum umbra-like virus 1 (SsUV1, KC601995). (E) Schematic representation of Botrytis cinerea deltaflexivirus 1 (BcDFV1), Botrytis cinerea mycotymovirus 1 (BcMTV1), and Botrytis cinerea flexivirus 1 (BcFlV1) RNA genomes showing the locations of ORFs. (F) Amino acid sequence alignment of mycoviruses tymo-like RdRp showing conserved motifs I to VIII, with the reference genomes of Sclerotinia sclerotiorum deltaflexivirus 2 (SsDFV2, MH299810) and Botrytis virus F (BVF, LN827953). (G) Percent identity matrix generated by Clustal Omega 2.1. Identities from higher to lower are labeled from dark green to dark red, respectively.