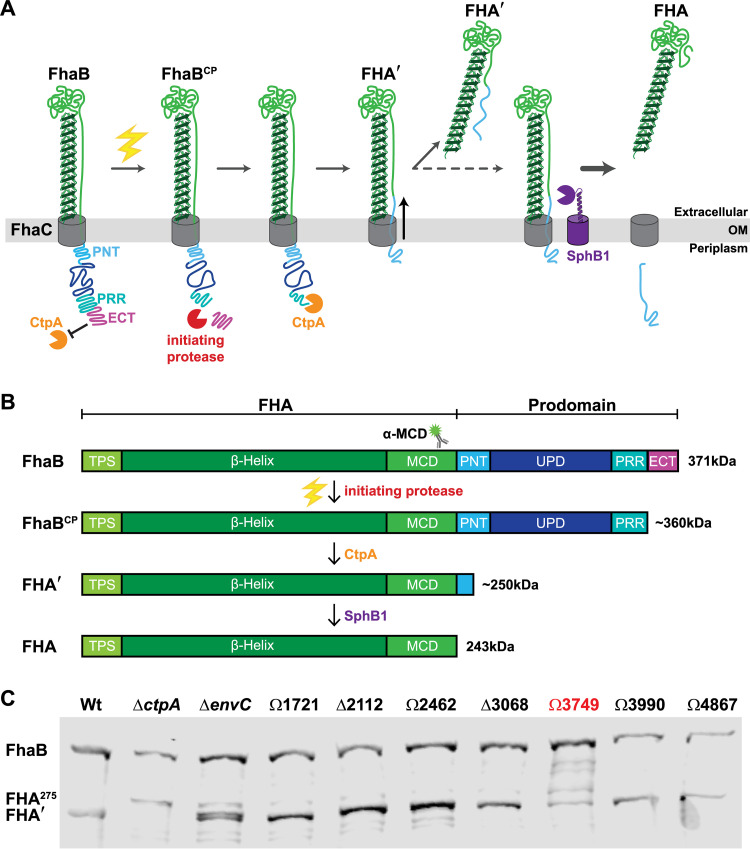
FIG 1.
A screen of FhaB processing in B. bronchiseptica strains that carry mutations of predicted extracytoplasmic proteases identifies BB3749 as encoding a putative FhaB protease. (A) Our current model of regulated FhaB processing by three proteases. An unknown signal (electric bolt) causes the initiating protease to remove the extreme C terminus (ECT) that acts as a guard against CtpA. This clipped product (FhaBCP) is recognized by CtpA, which degrades most of the prodomain to form FHA′. Removal of a portion of the PNT by CtpA causes a shift in the molecule that allows the release of FHA′ into the supernatant. Under certain conditions this shift also exposes the primary SphB1 cleavage site and allows SphB1 to create FHA (dashed arrow), which is immediately released. (B) Linear schematics of FhaB (without the extended signal sequence) and the main polypeptides that result from processing by each protease. Molecular weights (kDa) are listed on the right. (C) Western blot analysis of whole-cell lysates (WCL) from overnight cultures of B. bronchiseptica strains with deletions of (Δ) or plasmid disruptions in (Ω) the indicated ORFs. The antigenic region of the α-MCD polyclonal antibody is indicated by the cartoon of a fluorescent labeled antibody in panel B.