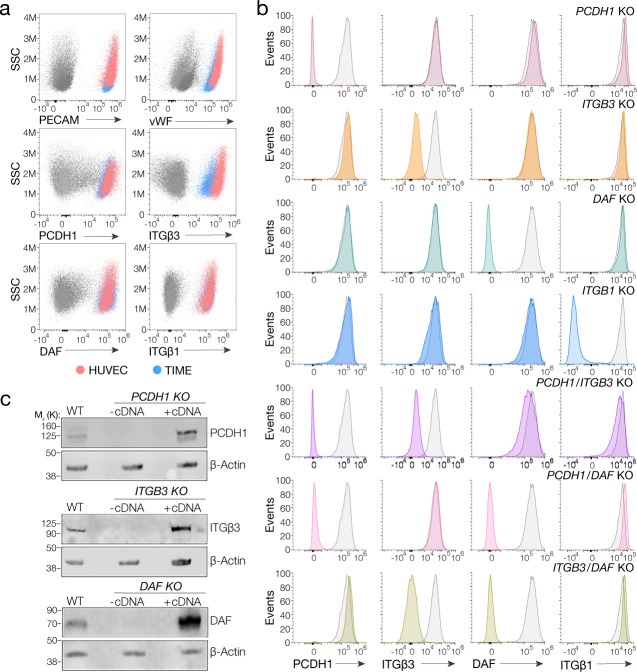
Figure 1. Suitability of TIME cells as a model to study hantavirus entry and the generation of knockout cells.
(a) Upper panels, total flow cytometry plots of HUVEC and TIME cells stained for endothelial cell markers PECAM and von Willebrand factor (vWF). Medium and lower panels, surface flow cytometry plots of HUVEC and TIME cells stained for PCDH1, β3 integrin, DAF, β1 integrin. (b) Surface flow cytometry of wild-type (WT) and knockout (KO) TIME cells stained as above. Histograms of WT cells are shown in gray; single- and double-KO cells are shown in color. (c) Western blot analysis of WT TIME cells and KO cells ± cDNA. β-Actin was used as a loading control.