Figure 1.
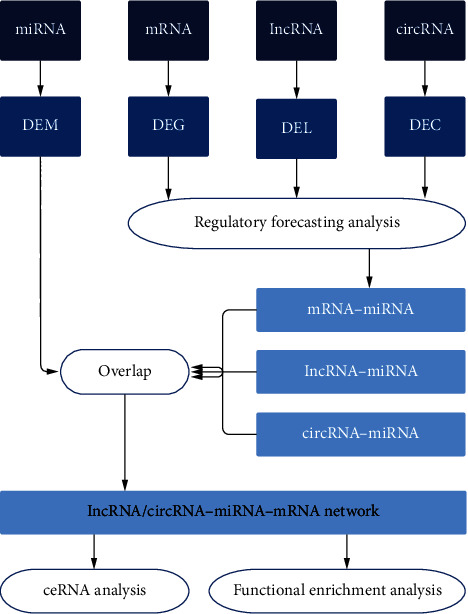
The logical framework is mainly divided into data collection and screening, data analysis, and network construction. The datasets of mRNA, miRNA, lncRNA, and circRNA correlated to IDD were collected from the public database, and DEMs, DEGs, DELs, and DECs with significant expression differences were screened out. Bioinformatics analysis tools and databases were used to conduct regulatory prediction analysis on DEGs, DELs, and DECs and obtain miRNAs paired with them. DEMs was then matched to establish a complete lncRNA/circRNA-miRNA-mRNA network. Then, the network was used to conduct ceRNA analysis and functional enrichment analysis. IDD: intervertebral disc degeneration; DEM: differentially expressed miRNA; DEG: differentially expressed mRNA; DEL: differentially expressed lncRNA; DEC: differentially expressed circRNA; ceRNA: competitive endogenous RNA.
