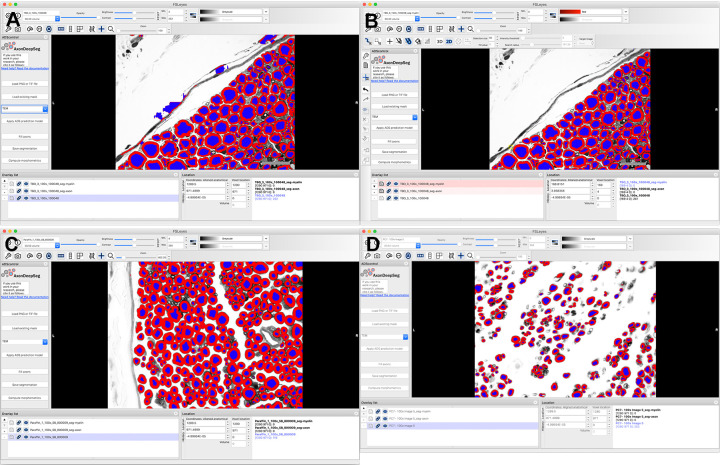
Fig 2. Representative screen shots showing user interface of AxonDeepSeg (ADS).
(A) .tiff images were imported individually into the program, resolution was defined, and transmission electron microscopy was chosen as it produces dark myelin and lighter axons, as in light microscopy. Segmentation was then performed automatically, creating different layers for the axons and myelin. (B) Minimal manual correction was performed to remove objects mislabeled as axons. Histomorphometric measurements and axon count were then calculated by pressing the “Compute morphometrics” button, and a .csv file with these values was generated and segmented images were saved. (A) and (B) are micrographs of naïve nerves prepared in toluidine-resin protocol. (C) Micrograph of naïve nerve, osmium-paraffin protocol. (D) Micrograph of regenerating nerve, osmium-paraffin protocol.