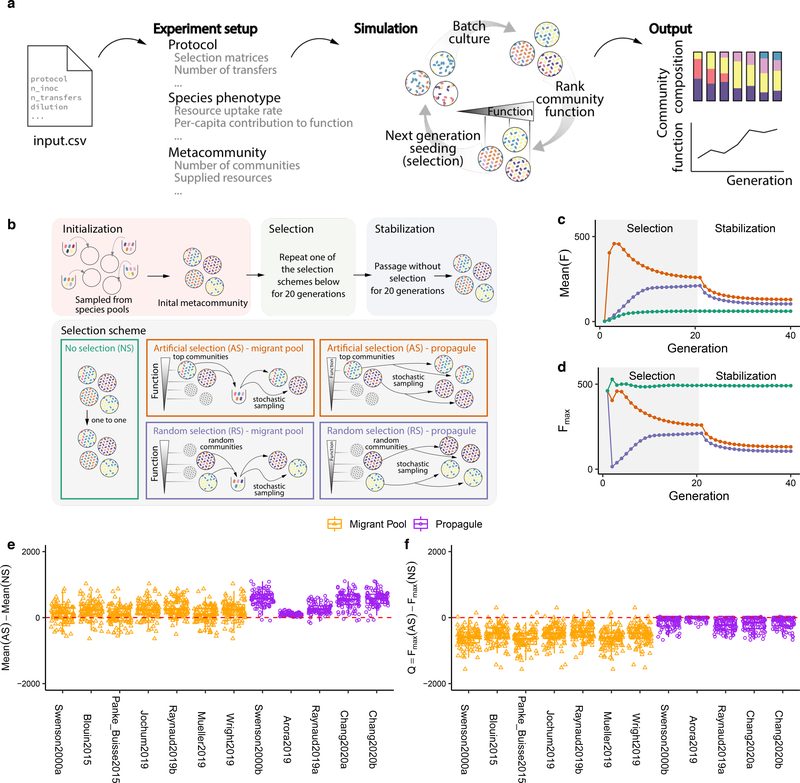
Figure 1. Migrant-pool and propagule strategies are limited in their ability to find new, high-functioning microbial communities.
(A) We constructed a Python package, ecoprospector, which allows us to artificially select arbitrarily large and diverse in silico communities. The experimental design of a selection protocol (e.g., number of communities, growth medium, method of artificial selection, function under selection, etc.) is entered in a single input .csv file (Methods). Communities are grown in serial batch-culture, where each transfer into a new habitat is referred to as a community “generation”. Within each batch incubation, species compete for nutrients from the supplied medium. At the end of the incubation period, communities are selected according to the specified, protocol-specific selection scheme, and the selected group is used to seed the communities in the offspring generation. Once the protocol is carried out to completion, ecoprospector outputs a simple text format for later analysis on community function and composition. (B) Illustration of previously used migrant pool and propagule selection schemes (AS) as well as the corresponding randomized controls (RS) 29,43. We also consider a no-selection ‘control’ scheme (NS). All protocols are applied at the end of each community generation and are implemented using a matrix representation depicted in Supplementary Fig. 1. A representative outcome of one community-level selection experiment is shown in (C-D), where we adapted the selection protocol from the migrant-pool strategy in ref 30. A metacommunity is seeded by inoculating ninoc = 106 randomly drawn cells from a species pool into each of 96 identical habitats and allowing them to grow (Methods). The metacommunity was then subject to 20 rounds of selection (generations), and then allowed to stabilize without selection for another 20 generations. The function maximized under selection F is additive on species contributions, whose per-capita species contribution to function is randomly generated (see main text). In each selection round, the top 20% communities with highest F (AS; red) (or a randomly chosen set in (RS; blue)) are selected and mixed into a single pool which is then used to seed all communities in the next generation by randomly sampling 106 cells into them. The NS protocol (green) simply propagates the communities in batch mode without selection. The changes in overall function over the generations is shown in (C) (average F) and (D) (maximum function Fmax). (E) Selection strategies were adapted from twelve experimental protocols in previous studies (see Supplementary Table 1; Methods). All were applied to standard metacommunity sizes (96 communities), for the same number of generations (20 selection generations + 20 stabilization generations). All protocols have a significantly greater mean function in the AS than in the NS line (two-sided paired t-test, P < 0.01) as well as the RS lines (Supplementary Fig. 4). (F) The difference in Fmax between the AS and NS lines (Q). All protocols show a Mean Q < 0 (two-sided Welch’s t-test, P < 0.01), indicating that they did not succeed at improving the function of the best stabilized community in the ancestral population.