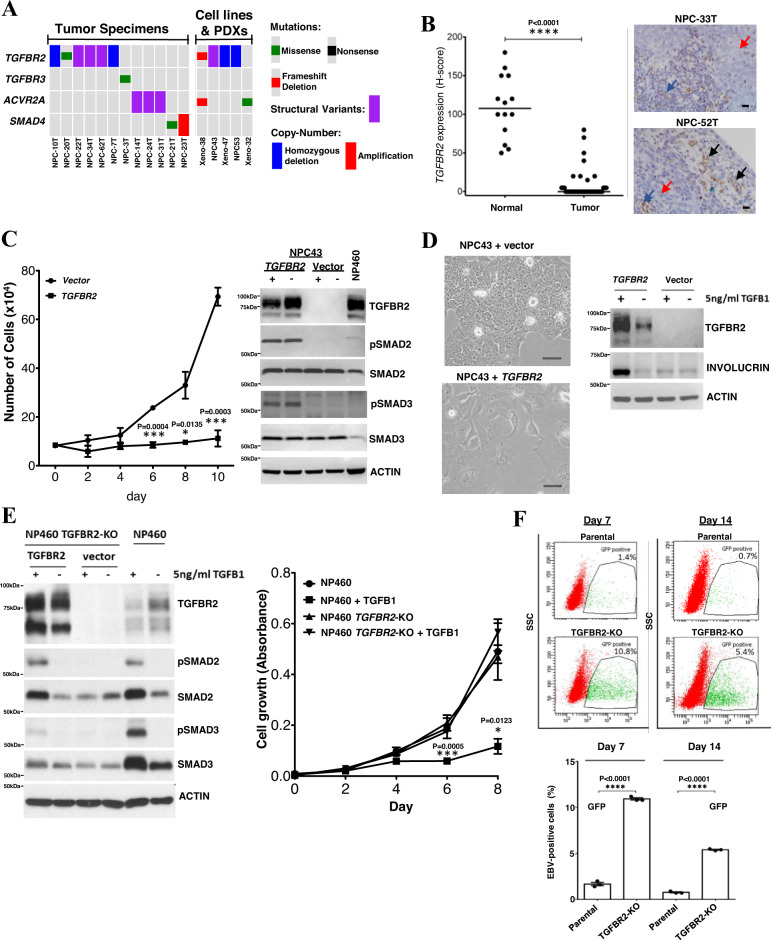
Fig. 5. TGFBR2 loss as a driver event in NPC.
A Somatic alterations of TGFBR2 and other genes in TGF-β/SMAD pathway. B RISH analysis of TGFBR2 expression in 41 NPCs and 14 adjacent normal epithelium. Significant downregulation of TGFBR2 was observed (Unpaired two-tailed t test, ****p < 0.0001, mean values of the data are presented) in NPC. Representative NPC cases (NPC33T, NPC-52T) showing no TGFBR2 expression in the tumor cells (red arrows) and positive signals in the infiltrating lymphocytes (blue arrows) and normal epithelium (black arrows). Scale bar: 10 μm. C Dramatic growth inhibition was found in wild-type TGFBR2 transfected TGFBR2-deleted NPC43 cells (unpaired two-tailed t test, *p < 0.05; ***p < 0.0005; data are presented as mean values ± SEM). Western blotting showed pSMAD2 and pSMAD3 overexpression in TGFBR2-expressing NPC43 cells. NP460 with TGFBR2 expression was included as control. Replication: n = 3 biologically independent experiments. D Re-introduction of wild-type TGFBR2 induced cell differentiation and morphological changes in NPC43 cells. In TGFBR2-expressing NPC43 cells, the levels of involucrin were increased upon TGF-β1 treatment, as compared with the vector control. Replication: n = 3 biologically independent experiments. Similar results were found in the repeated experiments. Scale bar: 50 μm. E CRISPR/Cas9-mediated knockout of TGFBR2 in NP460 (NP460KO) cells resulted in the loss of TGFBR2. No pSmad2 and pSmad3 expression were detected in NP460KO cells following TGF-β1 treatment. Re-expression of wild-type TGFBR2 in NP460KO cells restored its response to TGF-β1 as shown by the induction of pSmad2 and pSmad3. NP460KO cells showed significant growth inhibition when compared with parental NP460 cells after TGF-β1 treatment (unpaired two-tailed t test, *p < 0.05; ***p < 0.0005; data are presented as mean values ± SEM). Replication: n = 5 biologically independent experiments. F The parental NP460 and NP460KO cells were infected with a GFP-tagged recombinant EBV. After EBV infection and flow sorting of EBV-positive cells, significantly higher numbers of EBV-positive cells were maintained in TGFBR2 knockout NP460KO cells than those in parental cells on both day7 and day 14 (unpaired two-tailed t test, ****p < 0.0005, data are presented as mean values ± SEM). Replication: n = 3 biologically independent experiments. Source data are provided as a Source Data file.