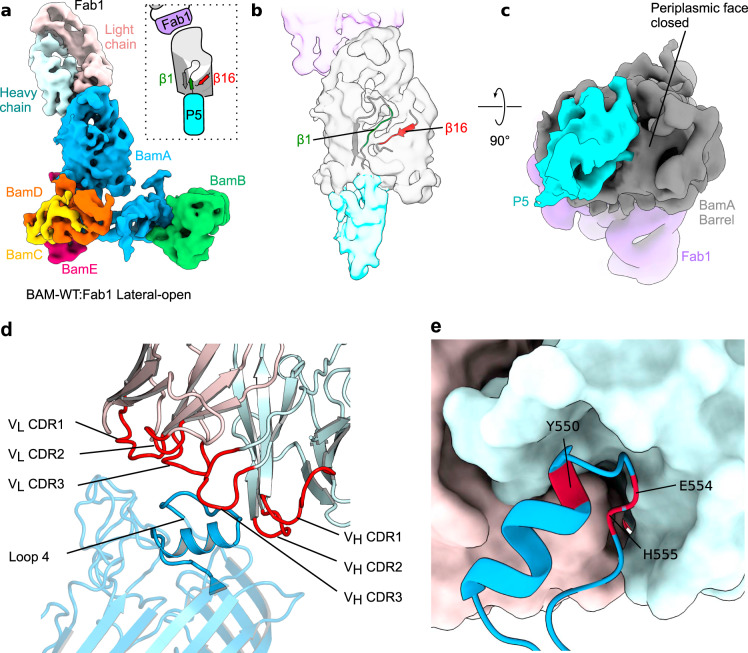
Fig. 3. Fab1-bound BAM is in a lateral-open conformation.
a 5.1 Å cryoEM map of the BAM–Fab1 complex in a lateral-open conformation at a contour of 10σ, coloured by subunit. The lateral gate is fully open and POTRA-5 occludes the BamA barrel (schematic inset). b Cartoon representation of the corresponding atomic model at the lateral gate superimposed on the segmented density for the barrel and POTRA-5 of BamA. β1 is in a conformation that makes limited contact with β16. c The same density viewed from the periplasmic side, showing that the BamA lumen is blocked by POTRA-5 in this conformation. Panels made using UCSF ChimeraX76. Segmenting and colouring performed with corresponding atomic models. Less well-resolved regions and the micelle have been masked. d Close up of the BamA–Fab1 interface region highlighting the Fab1 CDRs (red) interacting with eL4 of BamA (dark blue). Other regions of BamA are rendered semi-transparent to highlight eL4. Heavy and light chains of Fab1 are coloured cyan and pink, respectively. e The VL and VH domains of Fab1 variable form a complementary binding surface for eL4 of BamA involving residues Y550, E554 and H555 (red). Sidechains for Y550, E554 and H555 are not shown as they are not well-defined in the electron density, but the close location of these residues in the binding pocket is consistent with previous mutagenesis studies16.