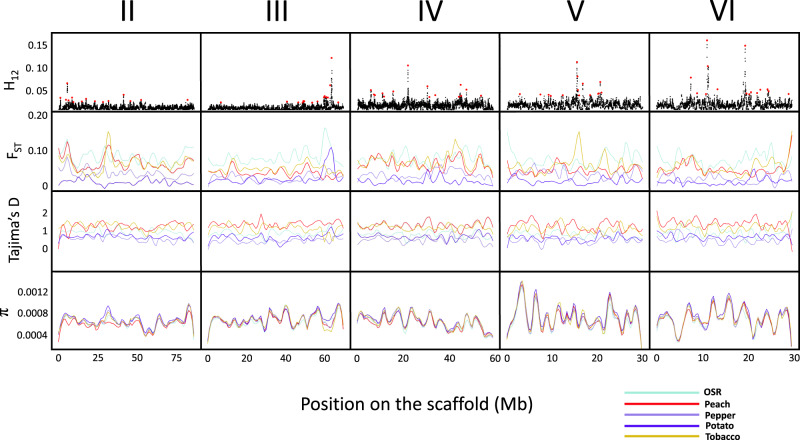
Fig. 4. Genomic divergence and signatures of selection associated with host plant use in M. persicae.
Panels from bottom to top display nucleotide diversity (π), Tajima’s D, FST, and H12 values across the 5 autosomal chromosomes of M. persicae for the main host plant groups (oilseed rape (OSR), peach, tobacco, pepper and potato), see Supplementary Data 1 for sample sizes. H12 scan: Each data point represents the H12 value calculated based on a 1000 SNP window. Red points highlight the top 15 peaks at each scaffold. Fixation index (FST), Tajima’s D, and nucleotide diversity (π): smoothed lines were estimated based on a 10 kb chromosomal window.