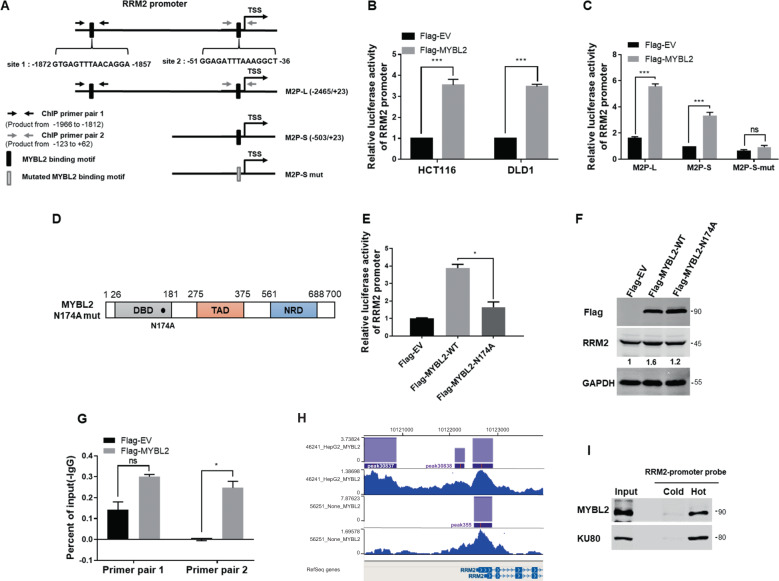
Fig. 3. MYBL2 activated the transcription of RRM2 by directly binding to RRM2 promoter in CRC cells.
A The schematic diagram for two MYBL2 biding motifs on RRM2 promoter and related truncated and site-mutated sequences. B Relative luciferase reporter activity of RRM2 promoter (−2465/+23) while co-transfected with MYBL2 expression plasmid for 48 h in HCT116 and DLD1 cells, respectively, pRL-SV40 as an internal control reporter. C Relative luciferase reporter activity of the truncated and mutated RRM2 promoters while co-transfected with MYBL2 expression plasmid for 48 h in DLD1 cells, pRL-SV40 as an internal control reporter. D The DNA-binding deficient mutation site in MYBL2 protein. E Relative luciferase reporter activity of RRM2 promoter (−2465/+23) while co-transfected with MYBL2 wild-type or N174A mutant. F The effects of overexpression of MYBL2 wild-type or N174A mutant on the expression of RRM2 in DLD1 cells. G Chromatin extracted from DLD1 cells was immunoprecipitated with the Flag or IgG antibodies, qRT-PCR were carried out on the immunoprecipitated DNAs using the specific primer pairs for the RRM2 promoter. H The ChIP-Seq data from Cistrome browser showed the binding of MYBL2 to RRM2 promoter in HepG2 cells. I Nuclear proteins obtained from DLD1 cells were pulled down by a non-biotin-labeled (cold) or biotin-labeled (hot) RRM2 promoter (−2465/+23) DNA probe, western blotting was used to detect the binding of MYBL2 or Ku80 (loading control). Unpaired Student’s t-test (2-tailed) was used to analyze the significance between different groups. ns, not significant; *p < 0.05; ***p < 0.001.