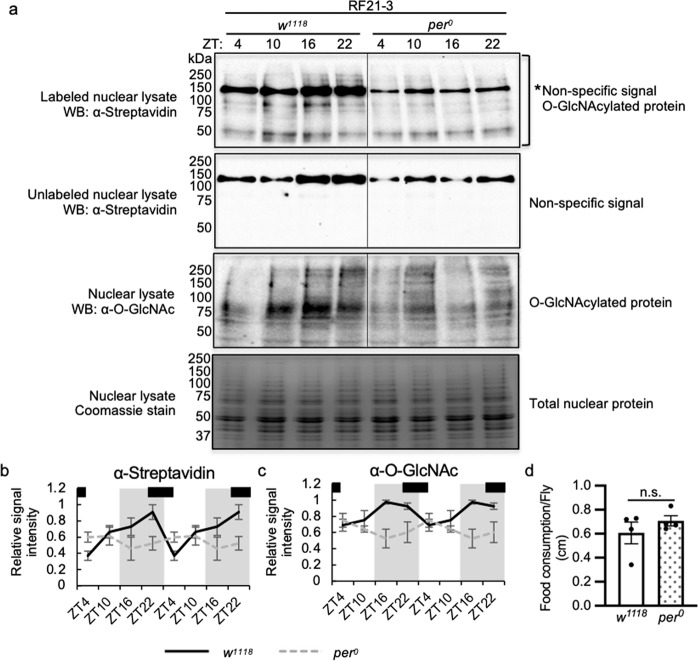
Fig. 6. The circadian clock is necessary to generate daily rhythms in protein O-GlcNAcylation.
a Western blots showing daily rhythms in nuclear protein O-GlcNAcylation in WT (w1118) and per0 flies fed between ZT21-3 (natural feeding time). Protein O-GlcNAcylation was detected using two methods: (top 2 panels) chemoenzymatic labeling in combination with immunoblotting with α-streptavidin and (third panel) immunoblotting with α-O-GlcNAc. Unlabeled samples (second panel) were processed in parallel to labeled samples (top panel) to identify non-specific signal. Total nuclear proteins stained by Coomassie blue (bottom) were used for normalization. The asterisk (top panel) denotes non-specific signal. Each biological replicate was performed independently with similar results (n = 4). b, c Quantification of protein O-GlcNAcylation detected using enzymatic labeling (n = 4; WT: p = 0.0096, per0: p = 0.44, RAIN; WT vs per0: p = 0.011, DODR), and using α-O-GlcNAc (n = 4; WT: p = 0.015, per0: p = 0.28, RAIN; WT vs per0: p = 0.017, DODR). Data are double plotted. Gray shading indicates the dark period. Black bars on the top of graphs denote the feeding periods for flies, ZT21-3. d Food consumption of WT and per0 flies over 24-hr period as measured by CAFE assay (n = 4; p = 0.35; two-tailed Student’s t-test). Data are presented as mean ± SEM.