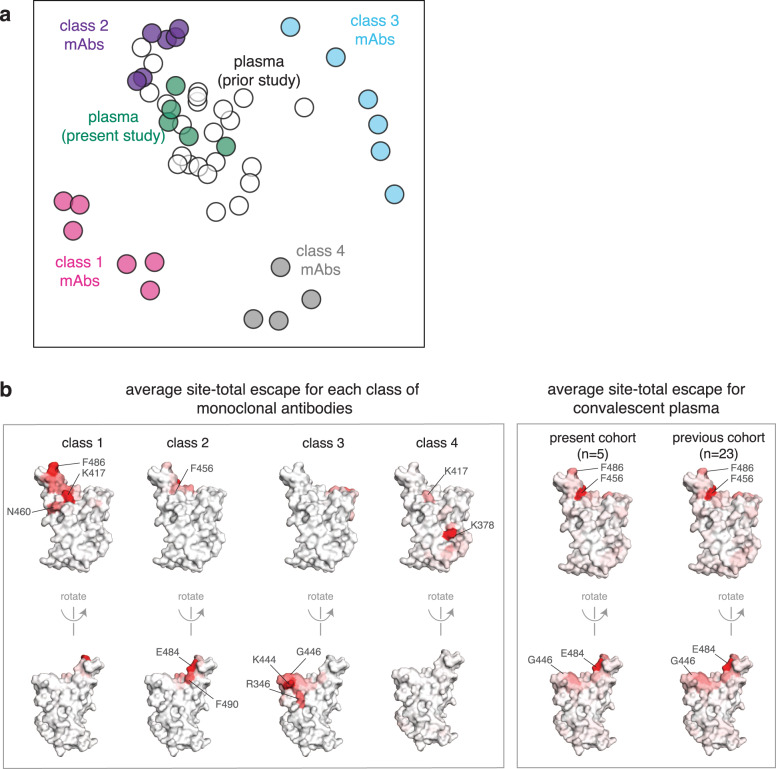
Fig. 4. The escape maps of convalescent polyclonal plasmas most resemble class 2 antibodies.
a Multidimensional scaling projection of the escape maps of polyclonal plasmas and monoclonal antibodies of each class. Antibodies or plasmas that are nearby in the plot have their binding affected by similar RBD mutations. The antibodies are those in Fig. 1c, colored according to antibody class, as in Fig. 1. The five plasmas newly mapped in this study are shown in green, and the previously mapped 23 plasmas18 are shown in white. b Structural projection of sites where mutations reduce binding by each class of monoclonal antibodies (left) or polyclonal plasmas (right). The RBD surface coloring is scaled from white to red, with white indicating no escape, and red indicating the site with the greatest average site-total escape for all antibodies or plasmas in that group. Mutations to sites such as E484, F456, and F486 have some of the largest effects on binding by polyclonal plasmas and class 2 antibodies. An interactive version of (a) that includes additional antibodies and vaccine sera is available at https://jbloomlab.github.io/SARS2_RBD_Ab_escape_maps/.