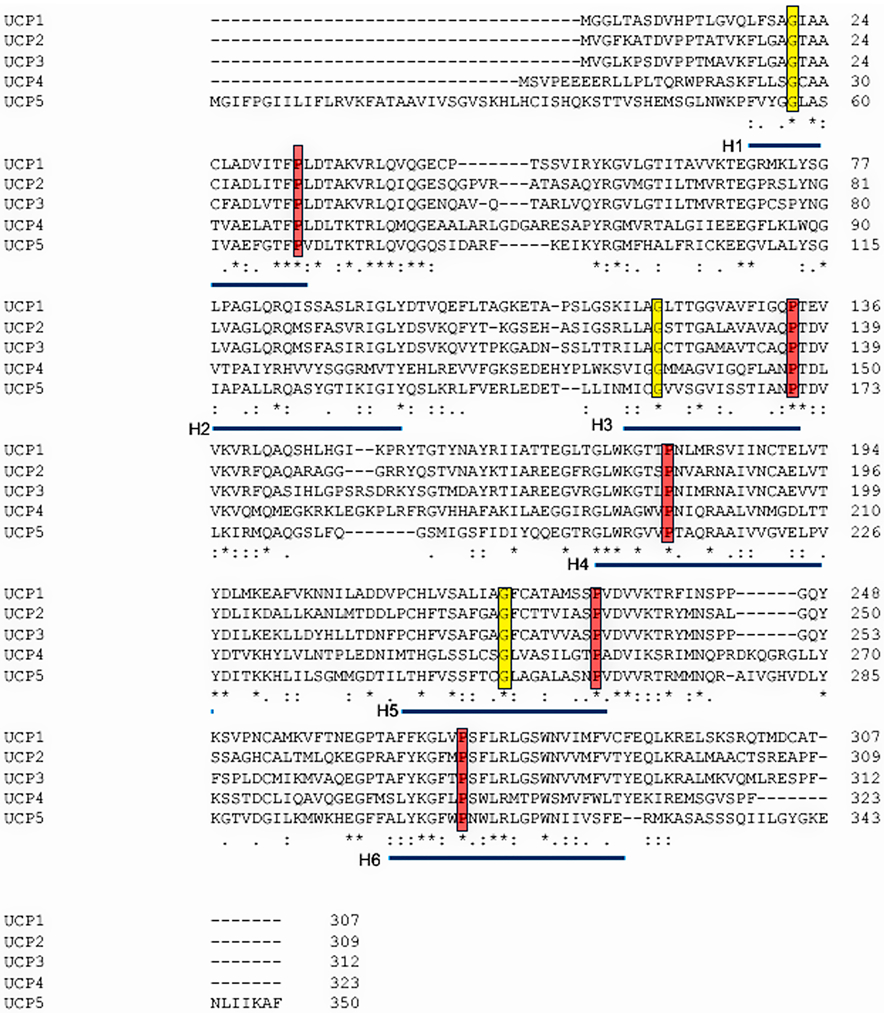
Figure 5. Sequence alignment of the five human UCP proteins.
Sequences used for alignment were the RefSeqs UCP1 (NP_068605.1), UCP2 (NP_003346.2), UCP3 (NP_003347.1), UCP4 (NP_004268.3) and UCP5 (XP_016885426.1). Alignment was carried out using the Clustal Omega program (Madeira et al., 2019). Conserved residues are marked with “*”, and conserved residues with “.” or “:”. The transmembrane helices are underlined in blue, key prolines in these helices outlined in red, and highly conserved glycines in yellow.