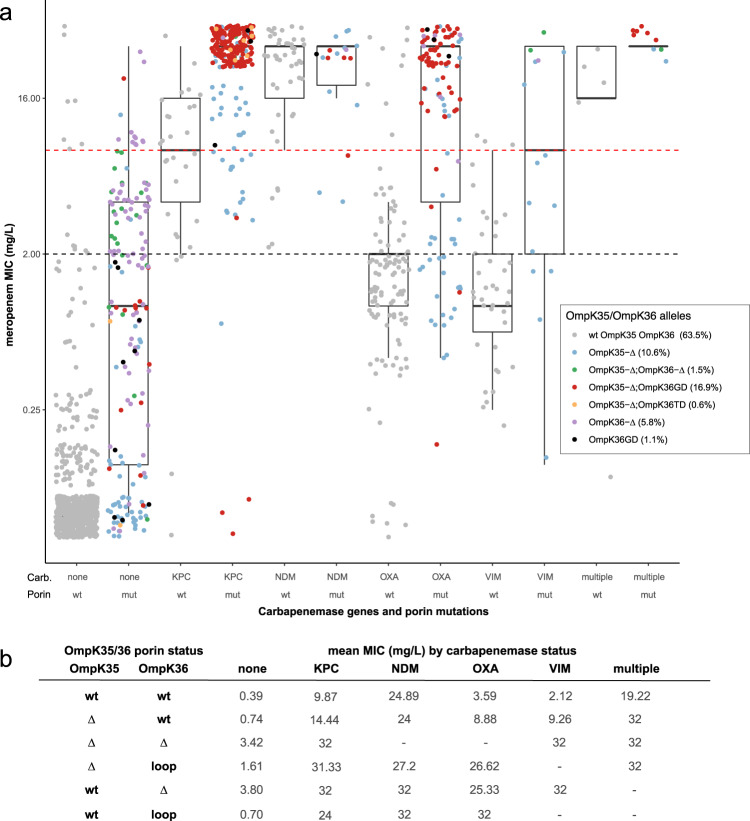
Fig. 3. Distribution of meropenem MIC, stratified by Kleborate-detected carbapenemase genes and OmpK35/36 porin mutations, for European K. pneumoniae surveillance isolates.
a Data shown summarize Kleborate results for 1490 K. pneumoniae genomes from the EuSCAPE study (data included in Supplementary Data 2). Each circle represents the reported meropenem MIC for an isolate, coloured by type of porin mutation/s identified by Kleborate from the corresponding genome assembly (colour key in inset legend, the prevalence of each genotype across 1490 genomes is indicated in brackets). Isolates are stratified by carbapenemase gene (enzymes labelled on x-axis) and OmpK mutations41, 42 reported by Kleborate. Wt, full-length OmpK35 and OmpK36 with no GD/TD insertion in the OmpK36 β-strand loop; mut, otherwise; Δ, missing/truncated. Dashed lines indicate EUCAST breakpoints for clinical resistance (red, MIC > 8) and non-susceptibility (black, MIC > 2). For each boxplot, the length of the box corresponds to the interquartile range with the centre line corresponding to the median, and the whiskers represent the minimum and maximum values. b Mean meropenem MIC values for the 1490 EuSCAPE isolates, grouped by the combination of porin gene status and the presence of carbapenemase genes. Porin status is expressed as: Δ, missing/truncated; loop, GD or TD insertion in the OmpK36 β-strand loop; wt, otherwise.