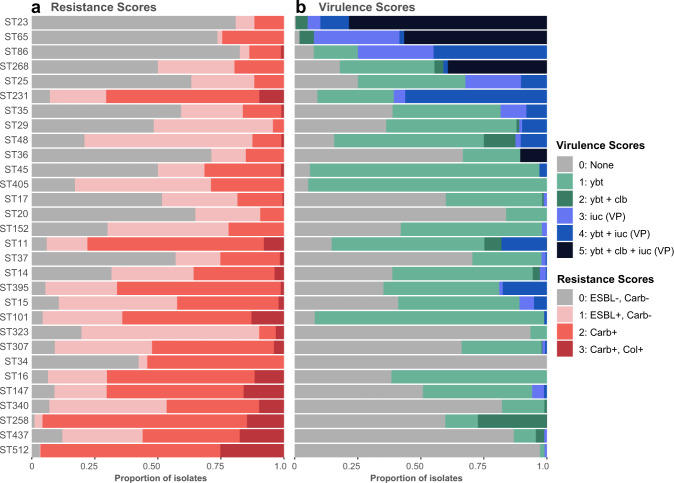
Fig. 5. Distribution of (a) resistance and (b) virulence scores among genomes belonging to the 30 most common K. pneumoniae lineages.
Data shown summarize Kleborate results for non-redundant set of 9705 publicly available K. pneumoniae genomes (Supplementary Data 2). Lineages were defined on the basis of multi-locus sequence types (STs) reported by Kleborate, and ordered from highest to lowest difference between mean virulence and mean resistance score. Minimum genome count per ST shown is 50. Ybt yersiniabactin, clb colibactin, iuc aerobactin, VP virulence plasmid, ESBL extended-spectrum β-lactamase, Carb carbapenemase, Col colistin resistance determinant.