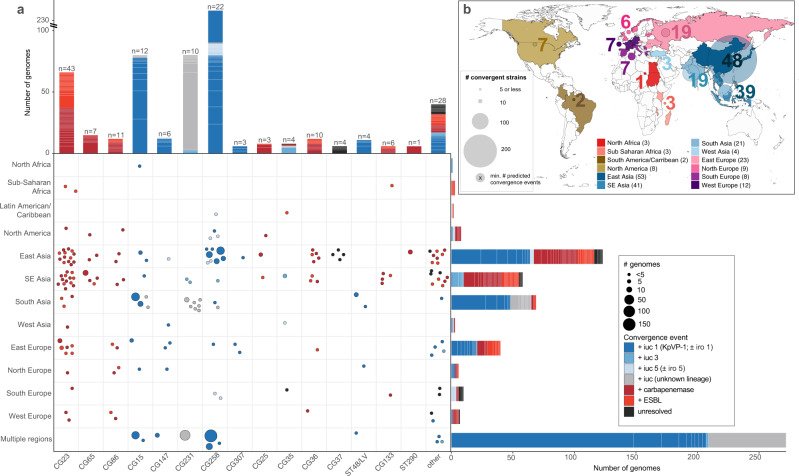
Fig. 7. Convergence of AMR and virulence determinants in the K. pneumoniae population, identified by Kleborate analysis of public genomes.
a Geographical and lineage distribution of convergence events. Each circle represents a unique convergence event (i.e. a monophyletic clade harbouring both ESBL/carbapenemase genes and iuc; see interactive tree at https://microreact.org/project/JDyan46yctyDh6weEUjWN, summary of events in Supplementary Data 7, assignment of genomes to events in Supplementary Data 2). Circles are scaled by the number of total genomes linked to the event and coloured to indicate whether convergence is inferred to have occurred via (i) acquisition of AMR gene/s (ESBL or carbapenemase/s) by a hypervirulent lineage, (ii) acquisition of an iuc-encoding plasmid by an AMR or non-AMR lineage, or (iii) unresolved means as per inset legend. Marginal barplots show the number of convergence events (colour blocks) and genomes (block heights) associated with each lineage (top) or geographical region (right). Lineages were defined on the basis of multi-locus sequence types (STs), number of convergence events estimated for each is labelled at the top of each bar. b Distribution of convergent genomes by location. Countries from which convergent genomes were detected are coloured on the map; circles represent the number of convergent genomes detected in each UN-defined geographical region (indicated by colour, as per inset legend), scaled and labelled with the minimum estimated number of unique convergence events specific to each region (excluding inter-regional convergence events). The total number of convergence events affecting each region, including region-specific and inter-regional convergence events, are given in brackets in the inset legend.