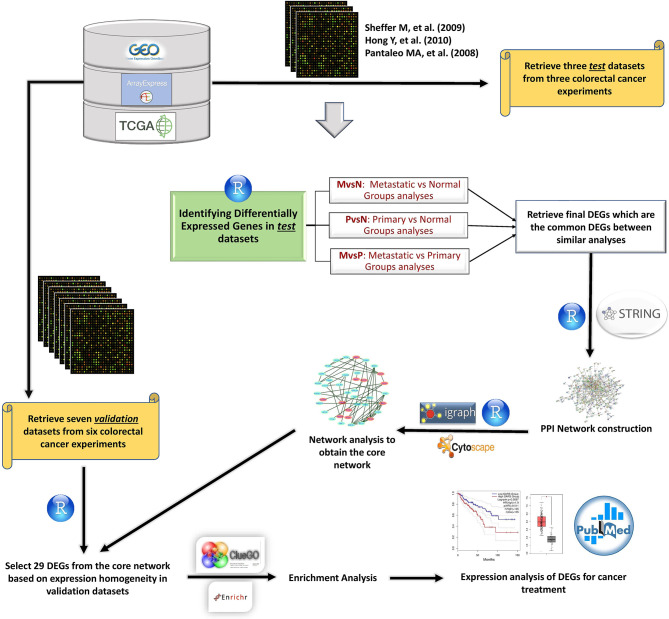
Figure 1.
The meta-analysis flowchart to attain therapeutic genetic targets. Gene expression datasets were extracted from different databases. Data were analyzed and visualized using R programming language. DEGs were obtained from analyzing Test datasets, then verified by Validation datasets. STRING database was utilized to construct the PPI network from DEGs. R software was used to analyze the network. Cytoscape was employed to visualize the networks, and enrichment results were obtained from ClueGO Cytoscape plugin and Enrichr online tools. Next, survival analysis and expression profiling were used for more validation of expression results. Finally, our results were compared to other studies, and molecular mechanism of validated DEGs was interrogated to propose a combination of target therapies.